Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway
- PMID: 19451544
- PMCID: PMC2704077
- DOI: 10.1261/rna.1541209
Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway
Abstract
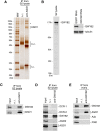
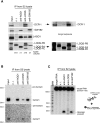
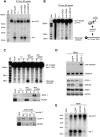
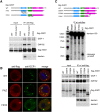
In Drosophila, miRNA is processed by Dicer-1 (DCR-1) from its precursor and loaded onto Argonaute1 (AGO1). AGO1 recognizes target mRNAs based on the miRNA sequence and suppresses the expression at post-transcriptional levels. GW182, a P-body component, localizes the AGO1 complex to processing bodies (P-bodies) where mRNA targets are decayed or stored. However, the details of the pathway remain elusive. In this study, two distinct types of AGO1-containing complexes from Drosophila Schneider2 (S2) cells were isolated and compared at the molecular level. The AGO1 complex with DCR-1 contained neither mature miRNA nor GW182 but exhibited pre-miRNA processing activity. The resultant mature RNA was loaded onto AGO1 within the complex. The AGO1 complex with GW182 excluded DCR-1, but possessed mature miRNA and showed no pre-miRNA processing activity. Thus, the AGO1-DCR-1 and AGO1-GW182 complexes correspond to miRLC (miRISC loading complex) and miRISC, respectively. The requirement for various domains of AGO1 in miRNA-loading and DCR-1/GW182 interaction was also examined. The Mid domain mutant (F2V2) interacted with DCR-1 but not with mature miRNA and GW182. The AGO1-PAZ mutant lacks the mature miRNA-binding ability but associates with either DCR-1 or GW182. The AGO1-PIWI mutant showed no Slicer activity but associates with mature miRNA. These results indicate that these domains are required differently for miRLC and miRISC formation in the miRNA pathway.
Figures

Similar articles
-
Structural determinants of miRNAs for RISC loading and slicer-independent unwinding.Nat Struct Mol Biol. 2009 Sep;16(9):953-60. doi: 10.1038/nsmb.1630. Epub 2009 Aug 16. Nat Struct Mol Biol. 2009. PMID: 19684602
-
R2D2 organizes small regulatory RNA pathways in Drosophila.Mol Cell Biol. 2011 Feb;31(4):884-96. doi: 10.1128/MCB.01141-10. Epub 2010 Dec 6. Mol Cell Biol. 2011. PMID: 21135122 Free PMC article.
-
The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1.Curr Biol. 2011 Nov 22;21(22):1878-87. doi: 10.1016/j.cub.2011.09.034. Epub 2011 Nov 3. Curr Biol. 2011. PMID: 22055293 Free PMC article.
-
siRNA and miRNA: an insight into RISCs.Trends Biochem Sci. 2005 Feb;30(2):106-14. doi: 10.1016/j.tibs.2004.12.007. Trends Biochem Sci. 2005. PMID: 15691656 Review.
-
The RNAi pathway initiated by Dicer-2 in Drosophila.Cold Spring Harb Symp Quant Biol. 2006;71:39-44. doi: 10.1101/sqb.2006.71.008. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381278 Review.
Cited by
-
Dicer is dispensable for asymmetric RISC loading in mammals.RNA. 2012 Jan;18(1):24-30. doi: 10.1261/rna.029785.111. Epub 2011 Nov 21. RNA. 2012. PMID: 22106413 Free PMC article.
-
Regulation of Argonaute slicer activity by guide RNA 3' end interactions with the N-terminal lobe.J Biol Chem. 2013 Mar 15;288(11):7829-7840. doi: 10.1074/jbc.M112.441030. Epub 2013 Jan 17. J Biol Chem. 2013. PMID: 23329841 Free PMC article.
-
Requirements for multivalent Yb body assembly in transposon silencing in Drosophila.EMBO Rep. 2019 Jul;20(7):e47708. doi: 10.15252/embr.201947708. Epub 2019 Apr 30. EMBO Rep. 2019. PMID: 31267711 Free PMC article.
-
Homeostatic control of Argonaute stability by microRNA availability.Nat Struct Mol Biol. 2013 Jul;20(7):789-95. doi: 10.1038/nsmb.2606. Epub 2013 May 26. Nat Struct Mol Biol. 2013. PMID: 23708604 Free PMC article.
-
Small RNA sorting: matchmaking for Argonautes.Nat Rev Genet. 2011 Jan;12(1):19-31. doi: 10.1038/nrg2916. Epub 2010 Nov 30. Nat Rev Genet. 2011. PMID: 21116305 Free PMC article. Review.
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Bushati N, Cohen SM. MicroRNA function. Annu Rev Cell Dev Biol. 2007;23:175–205. - PubMed
-
- Diederichs S, Haber DA. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
