Different members of a simple three-helix bundle protein family have very different folding rate constants and fold by different mechanisms
- PMID: 19445951
- PMCID: PMC2852649
- DOI: 10.1016/j.jmb.2009.05.010
Different members of a simple three-helix bundle protein family have very different folding rate constants and fold by different mechanisms
Abstract
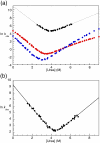
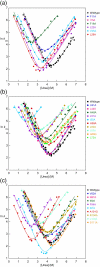
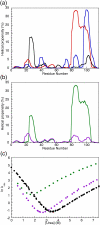
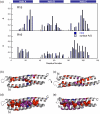
The 15th, 16th, and 17th repeats of chicken brain alpha-spectrin (R15, R16, and R17, respectively) are very similar in terms of structure and stability. However, R15 folds and unfolds 3 orders of magnitude faster than R16 and R17. This is unexpected. The rate-limiting transition state for R15 folding is investigated using protein engineering methods (Phi-value analysis) and compared with previously completed analyses of R16 and R17. Characterisation of many mutants suggests that all three proteins have similar complexity in the folding landscape. The early rate-limiting transition states of the three domains are similar in terms of overall structure, but there are significant differences in the patterns of Phi-values. R15 apparently folds via a nucleation-condensation mechanism, which involves concomitant folding and packing of the A- and C-helices, establishing the correct topology. R16 and R17 fold via a more framework-like mechanism, which may impede the search to find the correct packing of the helices, providing a possible explanation for the fast folding of R15.
Figures
Similar articles
-
Separating the effects of internal friction and transition state energy to explain the slow, frustrated folding of spectrin domains.Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):17795-9. doi: 10.1073/pnas.1201793109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711800 Free PMC article.
-
Protein folding: adding a nucleus to guide helix docking reduces landscape roughness.J Mol Biol. 2012 Oct 26;423(3):273-83. doi: 10.1016/j.jmb.2012.08.003. Epub 2012 Aug 20. J Mol Biol. 2012. PMID: 22917971 Free PMC article.
-
The folding of a family of three-helix bundle proteins: spectrin R15 has a robust folding nucleus, unlike its homologous neighbours.J Mol Biol. 2014 Apr 3;426(7):1600-10. doi: 10.1016/j.jmb.2013.12.018. Epub 2013 Dec 24. J Mol Biol. 2014. PMID: 24373753 Free PMC article.
-
From covalent transition states in chemistry to noncovalent in biology: from β- to Φ-value analysis of protein folding.Q Rev Biophys. 2024 Mar 20;57:e4. doi: 10.1017/S0033583523000045. Q Rev Biophys. 2024. PMID: 38597675 Review.
-
Morphogenesis of a protein: folding pathways and the energy landscape.Biochem Soc Trans. 2012 Apr;40(2):429-32. doi: 10.1042/BST20110683. Biochem Soc Trans. 2012. PMID: 22435825 Review.
Cited by
-
Separating the effects of internal friction and transition state energy to explain the slow, frustrated folding of spectrin domains.Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):17795-9. doi: 10.1073/pnas.1201793109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711800 Free PMC article.
-
Cooperative formation of native-like tertiary contacts in the ensemble of unfolded states of a four-helix protein.Proc Natl Acad Sci U S A. 2010 Jul 27;107(30):13306-11. doi: 10.1073/pnas.1003004107. Epub 2010 Jul 12. Proc Natl Acad Sci U S A. 2010. PMID: 20624986 Free PMC article.
-
Engineering a two-helix bundle protein for folding studies.Protein Eng Des Sel. 2010 May;23(5):357-64. doi: 10.1093/protein/gzp080. Epub 2010 Feb 3. Protein Eng Des Sel. 2010. PMID: 20130106 Free PMC article.
-
The denatured state dictates the topology of two proteins with almost identical sequence but different native structure and function.J Biol Chem. 2011 Feb 4;286(5):3863-72. doi: 10.1074/jbc.M110.155911. Epub 2010 Nov 29. J Biol Chem. 2011. PMID: 21118804 Free PMC article.
-
Polymer uncrossing and knotting in protein folding, and their role in minimal folding pathways.PLoS One. 2013;8(1):e53642. doi: 10.1371/journal.pone.0053642. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23365638 Free PMC article.
References
-
- Gunasekaran K., Eyles S.J., Hagler A.T., Gierasch L.M. Keeping it in the family: folding studies of related proteins. Curr. Opin. Struct. Biol. 2001;11:83–93. - PubMed
-
- Zarrine-Afsar A., Larson S.M., Davidson A.R. The family feud: do proteins with similar structures fold via the same pathway? J. Mol. Biol. 2005;15:42–49. - PubMed
-
- Religa T.L., Markson J.S., Mayor U., Freund S.M., Fersht A.R. Solution structure of a protein denatured state and folding intermediate. Nature. 2005;437:1053–1056. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

