The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition
- PMID: 19440361
- PMCID: PMC2679190
- DOI: 10.1371/journal.pone.0005571
The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition
Abstract
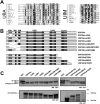
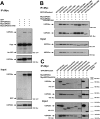
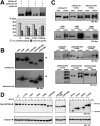
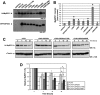
USP25m is the muscle isoform of the deubiquitinating (DUB) enzyme USP25. Similarly to most DUBs, data on USP25 regulation and substrate recognition is scarce. In silico analysis predicted three ubiquitin binding domains (UBDs) at the N-terminus: one ubiquitin-associated domain (UBA) and two ubiquitin-interacting motifs (UIMs), whereas no clear structural homology at the extended C-terminal region outside the catalytic domains were detected. In order to asses the contribution of the UBDs and the C-terminus to the regulation of USP25m catalytic activity, ubiquitination state and substrate interaction, serial and combinatorial deletions were generated. Our results showed that USP25m catalytic activity did not strictly depend on the UBDs, but required a coiled-coil stretch between amino acids 679 to 769. USP25 oligomerized but this interaction did not require either the UBDs or the C-terminus. Besides, USP25 was monoubiquitinated and able to autodeubiquitinate in a possible loop of autoregulation. UBDs favored the monoubiquitination of USP25m at the preferential site lysine 99 (K99). This residue had been previously shown to be a target for SUMO and this modification inhibited USP25 activity. We showed that mutation of K99 clearly diminished USP25-dependent rescue of the specific substrate MyBPC1 from proteasome degradation, thereby supporting a new mechanistic model, in which USP25m is regulated through alternative conjugation of ubiquitin (activating) or SUMO (inhibiting) to the same lysine residue (K99), which may promote the interaction with distinct intramolecular regulatory domains.
Conflict of interest statement
Figures

Similar articles
-
¹H, ¹³C and ¹⁵N backbone and side-chain resonance assignments of the N-terminal ubiquitin-binding domains of USP25.Biomol NMR Assign. 2014 Oct;8(2):255-8. doi: 10.1007/s12104-013-9495-1. Epub 2013 Jun 11. Biomol NMR Assign. 2014. PMID: 23754700
-
Tandem UIMs confer Lys48 ubiquitin chain substrate preference to deubiquitinase USP25.Sci Rep. 2017 Mar 22;7:45037. doi: 10.1038/srep45037. Sci Rep. 2017. PMID: 28327663 Free PMC article.
-
Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.Mol Cell. 2019 May 2;74(3):436-451.e7. doi: 10.1016/j.molcel.2019.02.030. Epub 2019 Mar 26. Mol Cell. 2019. PMID: 30926242 Free PMC article.
-
[Advances in the application of affinity separation for analyzing protein ubiquitination].Se Pu. 2021 Jan;39(1):26-33. doi: 10.3724/SP.J.1123.2020.07005. Se Pu. 2021. PMID: 34227356 Free PMC article. Review. Chinese.
-
Structure and function of the highly homologous deubiquitinases ubiquitin specific peptidase 25 and 28: Insights into their pathophysiological and therapeutic roles.Biochem Pharmacol. 2023 Jul;213:115624. doi: 10.1016/j.bcp.2023.115624. Epub 2023 May 26. Biochem Pharmacol. 2023. PMID: 37245535 Review.
Cited by
-
The ubiquitin interacting motifs of USP37 act on the proximal Ub of a di-Ub chain to enhance catalytic efficiency.Sci Rep. 2019 Mar 11;9(1):4119. doi: 10.1038/s41598-019-40815-z. Sci Rep. 2019. PMID: 30858488 Free PMC article.
-
Autologous K63 deubiquitylation within the BRCA1-A complex licenses DNA damage recognition.J Cell Biol. 2022 Sep 5;221(9):e202111050. doi: 10.1083/jcb.202111050. Epub 2022 Aug 8. J Cell Biol. 2022. PMID: 35938958 Free PMC article.
-
Deubiquitylation of deubiquitylases.Open Biol. 2017 Jun;7(6):170016. doi: 10.1098/rsob.170016. Open Biol. 2017. PMID: 28659380 Free PMC article. Review.
-
Ubiquitin-specific protease 25 functions in Endoplasmic Reticulum-associated degradation.PLoS One. 2012;7(5):e36542. doi: 10.1371/journal.pone.0036542. Epub 2012 May 9. PLoS One. 2012. PMID: 22590560 Free PMC article.
-
Deubiquitylase USP25 prevents degradation of BCR-ABL protein and ensures proliferation of Ph-positive leukemia cells.Oncogene. 2020 May;39(19):3867-3878. doi: 10.1038/s41388-020-1253-0. Epub 2020 Mar 12. Oncogene. 2020. PMID: 32203161
References
-
- Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182:1–11. - PubMed
-
- Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. - PubMed
-
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

