Studying the folding of multidomain proteins
- PMID: 19436439
- PMCID: PMC2645590
- DOI: 10.2976/1.2991513
Studying the folding of multidomain proteins
Abstract
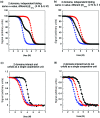
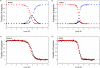
There have been relatively few detailed comprehensive studies of the folding of protein domains (or modules) in the context of their natural covalently linked neighbors. This is despite the fact that a significant proportion of the proteome consists of multidomain proteins. In this review we highlight some key experimental investigations of the folding of multidomain proteins to draw attention to the difficulties that can arise in analyzing such systems. The evidence suggests that interdomain interactions can significantly affect stability, folding, and unfolding rates. However, preliminary studies suggest that folding pathways are unaffected-to this extent domains can be truly considered to be independent folding units. Nonetheless, it is clear that interactions between domains cannot be ignored, in particular when considering the effects of mutations.
Figures


Similar articles
-
Distinguishing specific and nonspecific interdomain interactions in multidomain proteins.Biophys J. 2008 Jan 15;94(2):622-8. doi: 10.1529/biophysj.107.119123. Epub 2007 Sep 21. Biophys J. 2008. PMID: 17890397 Free PMC article.
-
Apparent cooperativity in the folding of multidomain proteins depends on the relative rates of folding of the constituent domains.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18113-8. doi: 10.1073/pnas.0604580103. Epub 2006 Nov 15. Proc Natl Acad Sci U S A. 2006. PMID: 17108086 Free PMC article.
-
Understanding the molecular basis of folding cooperativity through a comparative analysis of a multidomain protein and its isolated domains.J Biol Chem. 2023 Mar;299(3):102983. doi: 10.1016/j.jbc.2023.102983. Epub 2023 Feb 3. J Biol Chem. 2023. PMID: 36739950 Free PMC article.
-
The folding and evolution of multidomain proteins.Nat Rev Mol Cell Biol. 2007 Apr;8(4):319-30. doi: 10.1038/nrm2144. Epub 2007 Mar 14. Nat Rev Mol Cell Biol. 2007. PMID: 17356578 Review.
-
Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water.Adv Biochem Eng Biotechnol. 1998;61:37-85. doi: 10.1007/BFb0102289. Adv Biochem Eng Biotechnol. 1998. PMID: 9670797 Review.
Cited by
-
Personalized biochemistry and biophysics.Biochemistry. 2015 Apr 28;54(16):2551-9. doi: 10.1021/acs.biochem.5b00189. Epub 2015 Apr 15. Biochemistry. 2015. PMID: 25856502 Free PMC article.
-
Two immunoglobulin tandem proteins with a linking β-strand reveal unexpected differences in cooperativity and folding pathways.J Mol Biol. 2012 Feb 10;416(1):137-47. doi: 10.1016/j.jmb.2011.12.012. Epub 2011 Dec 13. J Mol Biol. 2012. PMID: 22197372 Free PMC article.
-
Domain tethering impacts dimerization and DNA-mediated allostery in the human transcription factor FoxP1.J Chem Phys. 2023 May 21;158(19):195101. doi: 10.1063/5.0138782. J Chem Phys. 2023. PMID: 37184020 Free PMC article.
-
A novel ulvan lyase family with broad-spectrum activity from the ulvan utilisation loci of Formosa agariphila KMM 3901.Sci Rep. 2018 Oct 2;8(1):14713. doi: 10.1038/s41598-018-32922-0. Sci Rep. 2018. PMID: 30279430 Free PMC article.
-
Plasticity of the proteasome-targeting signal Fat10 enhances substrate degradation.Elife. 2024 Jul 10;13:e91122. doi: 10.7554/eLife.91122. Elife. 2024. PMID: 38984715 Free PMC article.
References
-
- Apic, G, Gough, J, and Teichmann, S A (2001). “Domain combinations in archaeal, eubacterial and eukaryotic proteomes.” J. Mol. Biol. JMOBAK 310, 311–325. - PubMed
-
- Arora, P, Hammes, G G, and Oas, T G (2006). “Folding mechanism of a multiple-independently folding domain protein: double, B. domain of protein A.” Biochemistry BICHAW 45, 12312–12324. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
