TOPCONS: consensus prediction of membrane protein topology
- PMID: 19429891
- PMCID: PMC2703981
- DOI: 10.1093/nar/gkp363
TOPCONS: consensus prediction of membrane protein topology
Abstract
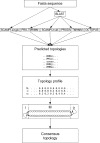
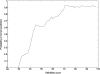
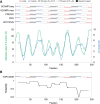
TOPCONS (http://topcons.net/) is a web server for consensus prediction of membrane protein topology. The underlying algorithm combines an arbitrary number of topology predictions into one consensus prediction and quantifies the reliability of the prediction based on the level of agreement between the underlying methods, both on the protein level and on the level of individual TM regions. Benchmarking the method shows that overall performance levels match the best available topology prediction methods, and for sequences with high reliability scores, performance is increased by approximately 10 percentage points. The web interface allows for constraining parts of the sequence to a known inside/outside location, and detailed results are displayed both graphically and in text format.
Figures
Similar articles
-
The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides.Nucleic Acids Res. 2015 Jul 1;43(W1):W401-7. doi: 10.1093/nar/gkv485. Epub 2015 May 12. Nucleic Acids Res. 2015. PMID: 25969446 Free PMC article.
-
Rapid membrane protein topology prediction.Bioinformatics. 2011 May 1;27(9):1322-3. doi: 10.1093/bioinformatics/btr119. Bioinformatics. 2011. PMID: 21493661 Free PMC article.
-
ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W390-3. doi: 10.1093/nar/gkh380. Nucleic Acids Res. 2004. PMID: 15215417 Free PMC article.
-
CCTOP: a Consensus Constrained TOPology prediction web server.Nucleic Acids Res. 2015 Jul 1;43(W1):W408-12. doi: 10.1093/nar/gkv451. Epub 2015 May 5. Nucleic Acids Res. 2015. PMID: 25943549 Free PMC article.
-
Topology of membrane proteins-predictions, limitations and variations.Curr Opin Struct Biol. 2018 Jun;50:9-17. doi: 10.1016/j.sbi.2017.10.003. Epub 2017 Nov 5. Curr Opin Struct Biol. 2018. PMID: 29100082 Review.
Cited by
-
Identification and analysis of cation channel homologues in human pathogenic fungi.PLoS One. 2012;7(8):e42404. doi: 10.1371/journal.pone.0042404. Epub 2012 Aug 2. PLoS One. 2012. PMID: 22876320 Free PMC article.
-
SpyA is a membrane-bound ADP-ribosyltransferase of Streptococcus pyogenes which modifies a streptococcal peptide, SpyB.Mol Microbiol. 2012 Mar;83(5):936-52. doi: 10.1111/j.1365-2958.2012.07979.x. Epub 2012 Jan 30. Mol Microbiol. 2012. PMID: 22288436 Free PMC article.
-
Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase.J Biol Chem. 2013 Nov 8;288(45):32211-32228. doi: 10.1074/jbc.M113.510313. Epub 2013 Sep 17. J Biol Chem. 2013. PMID: 24045953 Free PMC article.
-
Identification of intracellular and plasma membrane calcium channel homologues in pathogenic parasites.PLoS One. 2011;6(10):e26218. doi: 10.1371/journal.pone.0026218. Epub 2011 Oct 14. PLoS One. 2011. PMID: 22022573 Free PMC article.
-
The α-proteobacteria Wolbachia pipientis protein disulfide machinery has a regulatory mechanism absent in γ-proteobacteria.PLoS One. 2013 Nov 25;8(11):e81440. doi: 10.1371/journal.pone.0081440. eCollection 2013. PLoS One. 2013. PMID: 24282596 Free PMC article.
References
-
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. - PubMed
-
- Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. - PubMed
-
- Nilsson J, Persson B, von Heijne G. Consensus predictions of membrane protein topology. FEBS Lett. 2000;486:267–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

