Replication fork reversal and the maintenance of genome stability
- PMID: 19406929
- PMCID: PMC2699526
- DOI: 10.1093/nar/gkp244
Replication fork reversal and the maintenance of genome stability
Abstract
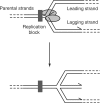
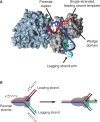
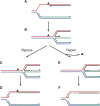
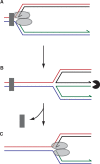
The progress of replication forks is often threatened in vivo, both by DNA damage and by proteins bound to the template. Blocked forks must somehow be restarted, and the original blockage cleared, in order to complete genome duplication, implying that blocked fork processing may be critical for genome stability. One possible pathway that might allow processing and restart of blocked forks, replication fork reversal, involves the unwinding of blocked forks to form four-stranded structures resembling Holliday junctions. This concept has gained increasing popularity recently based on the ability of such processing to explain many genetic observations, the detection of unwound fork structures in vivo and the identification of enzymes that have the capacity to catalyse fork regression in vitro. Here, we discuss the contexts in which fork regression might occur, the factors that may promote such a reaction and the possible roles of replication fork unwinding in normal DNA metabolism.
Figures


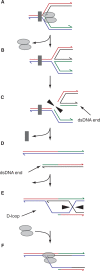
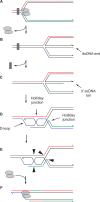
Similar articles
-
Mycobacterium tuberculosis RecG protein but not RuvAB or RecA protein is efficient at remodeling the stalled replication forks: implications for multiple mechanisms of replication restart in mycobacteria.J Biol Chem. 2015 Oct 2;290(40):24119-39. doi: 10.1074/jbc.M115.671164. Epub 2015 Aug 14. J Biol Chem. 2015. PMID: 26276393 Free PMC article.
-
Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8227-34. doi: 10.1073/pnas.111008698. Proc Natl Acad Sci U S A. 2001. PMID: 11459957 Free PMC article.
-
Time for remodeling: SNF2-family DNA translocases in replication fork metabolism and human disease.DNA Repair (Amst). 2020 Nov;95:102943. doi: 10.1016/j.dnarep.2020.102943. Epub 2020 Aug 15. DNA Repair (Amst). 2020. PMID: 32971328 Free PMC article. Review.
-
Action of RuvAB at replication fork structures.J Biol Chem. 2001 Nov 9;276(45):41938-44. doi: 10.1074/jbc.M107945200. Epub 2001 Sep 10. J Biol Chem. 2001. PMID: 11551967
-
Mechanisms and regulation of replication fork reversal.DNA Repair (Amst). 2024 Sep;141:103731. doi: 10.1016/j.dnarep.2024.103731. Epub 2024 Jul 22. DNA Repair (Amst). 2024. PMID: 39089193 Review.
Cited by
-
Rescuing stalled or damaged replication forks.Cold Spring Harb Perspect Biol. 2013 May 1;5(5):a012815. doi: 10.1101/cshperspect.a012815. Cold Spring Harb Perspect Biol. 2013. PMID: 23637285 Free PMC article. Review.
-
Embracing Heterogeneity: Challenging the Paradigm of Replisomes as Deterministic Machines.Chem Rev. 2023 Dec 13;123(23):13419-13440. doi: 10.1021/acs.chemrev.3c00436. Epub 2023 Nov 16. Chem Rev. 2023. PMID: 37971892 Free PMC article. Review.
-
FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress.Nat Commun. 2013;4:1423. doi: 10.1038/ncomms2395. Nat Commun. 2013. PMID: 23361013
-
DisA Limits RecG Activities at Stalled or Reversed Replication Forks.Cells. 2021 May 31;10(6):1357. doi: 10.3390/cells10061357. Cells. 2021. PMID: 34073022 Free PMC article.
-
Role of Cockayne Syndrome Group B Protein in Replication Stress: Implications for Cancer Therapy.Int J Mol Sci. 2022 Sep 6;23(18):10212. doi: 10.3390/ijms231810212. Int J Mol Sci. 2022. PMID: 36142121 Free PMC article. Review.
References
-
- Setlow RB, Swenson PA, Carrier WL. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142:1464–1466. - PubMed
-
- Hanawalt PC. The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem. Photobiol. 1966;5:1–12. - PubMed
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976;101:417–425. - PubMed
-
- Fujiwara Y, Tatsumi M. Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: caffeine sensitive and caffeine resistant mechanisms. Mutat. Res. 1976;37:91–110. - PubMed

