An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types
- PMID: 19399471
- PMCID: PMC2688022
- DOI: 10.1007/s00109-009-0467-y
An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types
Abstract
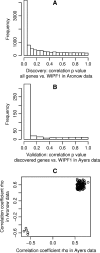
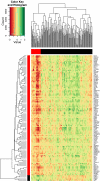
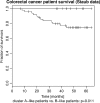
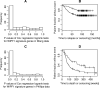
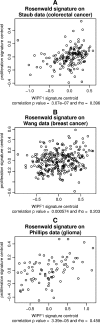
Wiskott-Aldrich syndrome (WAS) predisposes patients to leukemia and lymphoma. WAS is caused by mutations in the protein WASP which impair its interaction with the WIPF1 protein. Here, we aim to identify a module of WIPF1-coexpressed genes and to assess its use as a prognostic signature for colorectal cancer, glioma, and breast cancer patients. Two public colorectal cancer microarray data sets were used for discovery and validation of the WIPF1 co-expression module. Based on expression of the WIPF1 signature, we classified more than 400 additional tumors with microarray data from our own experiments or from publicly available data sets according to their WIPF1 signature expression. This allowed us to separate patient populations for colorectal cancers, breast cancers, and gliomas for which clinical characteristics like survival times and times to relapse were analyzed. Groups of colorectal cancer, breast cancer, and glioma patients with low expression of the WIPF1 co-expression module generally had a favorable prognosis. In addition, the majority of WIPF1 signature genes are individually correlated with disease outcome in different studies. Literature gene network analysis revealed that among WIPF1 co-expressed genes known direct transcriptional targets of c-myc, ESR1 and p53 are enriched. The mean expression profile of WIPF1 signature genes is correlated with the profile of a proliferation signature. The WIPF1 signature is the first microarray-based prognostic expression signature primarily developed for colorectal cancer that is instrumental in other tumor types: low expression of the WIPF1 module is associated with better prognosis.
Figures

Similar articles
-
Epigenetically upregulated WIPF1 plays a major role in BRAF V600E-promoted papillary thyroid cancer aggressiveness.Oncotarget. 2017 Jan 3;8(1):900-914. doi: 10.18632/oncotarget.13400. Oncotarget. 2017. PMID: 27863429 Free PMC article.
-
WIPF1 antagonizes the tumor suppressive effect of miR-141/200c and is associated with poor survival in patients with PDAC.J Exp Clin Cancer Res. 2018 Jul 24;37(1):167. doi: 10.1186/s13046-018-0848-6. J Exp Clin Cancer Res. 2018. PMID: 30041660 Free PMC article.
-
Systematic functional perturbations uncover a prognostic genetic network driving human breast cancer.Oncotarget. 2017 Mar 15;8(13):20572-20587. doi: 10.18632/oncotarget.16244. Oncotarget. 2017. PMID: 28411283 Free PMC article.
-
A gene expression signature identifies two prognostic subgroups of basal breast cancer.Breast Cancer Res Treat. 2011 Apr;126(2):407-20. doi: 10.1007/s10549-010-0897-9. Epub 2010 May 21. Breast Cancer Res Treat. 2011. PMID: 20490655
-
Evaluation of public cancer datasets and signatures identifies TP53 mutant signatures with robust prognostic and predictive value.BMC Cancer. 2015 Mar 26;15:179. doi: 10.1186/s12885-015-1102-7. BMC Cancer. 2015. PMID: 25886164 Free PMC article.
Cited by
-
In silico and Ex vivo approaches identify a role for toll-like receptor 4 in colorectal cancer.J Exp Clin Cancer Res. 2014 May 22;33(1):45. doi: 10.1186/1756-9966-33-45. J Exp Clin Cancer Res. 2014. PMID: 24887394 Free PMC article.
-
NQO1 inhibits proteasome-mediated degradation of HIF-1α.Nat Commun. 2016 Dec 14;7:13593. doi: 10.1038/ncomms13593. Nat Commun. 2016. PMID: 27966538 Free PMC article.
-
PML promotes metastasis of triple-negative breast cancer through transcriptional regulation of HIF1A target genes.JCI Insight. 2017 Feb 23;2(4):e87380. doi: 10.1172/jci.insight.87380. JCI Insight. 2017. PMID: 28239645 Free PMC article.
-
Revisiting the Concept of Stress in the Prognosis of Solid Tumors: A Role for Stress Granules Proteins?Cancers (Basel). 2020 Sep 1;12(9):2470. doi: 10.3390/cancers12092470. Cancers (Basel). 2020. PMID: 32882814 Free PMC article. Review.
-
Epigenetically upregulated WIPF1 plays a major role in BRAF V600E-promoted papillary thyroid cancer aggressiveness.Oncotarget. 2017 Jan 3;8(1):900-914. doi: 10.18632/oncotarget.13400. Oncotarget. 2017. PMID: 27863429 Free PMC article.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '10878810', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10878810/'}]}
- Moreau V, Frischknecht F, Reckmann I et al (2002) A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol 2:441–448 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1073/pnas.94.26.14671', 'is_inner': False, 'url': 'https://doi.org/10.1073/pnas.94.26.14671'}, {'type': 'PMC', 'value': 'PMC25088', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC25088/'}, {'type': 'PubMed', 'value': '9405671', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9405671/'}]}
- Ramesh N, Antón IM, Hartwig JH et al (1997) WIP, a protein associated with Wiskott–Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci U S A 94:14671–14676 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S0092-8674(02)01076-0', 'is_inner': False, 'url': 'https://doi.org/10.1016/s0092-8674(02)01076-0'}, {'type': 'PubMed', 'value': '12437929', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12437929/'}]}
- Volkman BF, Prehoda KE, Scott JA et al (2002) Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott–Aldrich Syndrome. Cell 111:565–576 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1074/jbc.M609902200', 'is_inner': False, 'url': 'https://doi.org/10.1074/jbc.m609902200'}, {'type': 'PubMed', 'value': '17229736', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17229736/'}]}
- Peterson FC, Deng Q, Zettl M et al (2007) Multiple WASP-interacting protein recognition motifs are required for a functional interaction with N-WASP. J Biol Chem 282:8446–8453 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1002/path.2088', 'is_inner': False, 'url': 'https://doi.org/10.1002/path.2088'}, {'type': 'PubMed', 'value': '17086554', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17086554/'}]}
- Curcio C, Pannellini T, Lanzardo S et al (2007) WIP null mice display a progressive immunological disorder that resembles Wiskott–Aldrich syndrome. J Pathol 211:67–75 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

