An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing
- PMID: 19394293
- PMCID: PMC2756231
- DOI: 10.1016/j.molcel.2009.02.032
An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing
Abstract
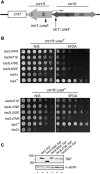
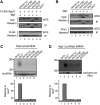
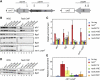
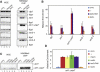
RNA interference (RNAi) plays a pivotal role in the formation of heterochromatin at the fission yeast centromeres. The RNA-induced transcriptional silencing (RITS) complex, composed of heterochromatic small interfering RNAs (siRNAs), the siRNA-binding protein Ago1, the chromodomain protein Chp1, and the Ago1/Chp1-interacting protein Tas3, provides a physical tether between the RNAi and heterochromatin assembly pathways. Here, we report the structural and functional characterization of a C-terminal Tas3 alpha-helical motif (TAM), which self-associates into a helical polymer and is required for cis spreading of RITS in centromeric DNA regions. Site-directed mutations of key residues within the hydrophobic monomer-monomer interface disrupt Tas3-TAM polymeric self-association in vitro and result in loss of gene silencing, spreading of RITS, and a dramatic reduction in centromeric siRNAs in vivo. These results demonstrate that, in addition to the chromodomain of Chp1 and siRNA-loaded Ago1, Tas3 self-association is required for RITS spreading and efficient heterochromatic gene silencing at centromeric repeat regions.
Figures


Similar articles
-
RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article.
-
Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin.Mol Cell Biol. 2008 Apr;28(7):2154-66. doi: 10.1128/MCB.01637-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212052 Free PMC article.
-
Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex.PLoS Genet. 2010 Oct 28;6(10):e1001174. doi: 10.1371/journal.pgen.1001174. PLoS Genet. 2010. PMID: 21060862 Free PMC article.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
-
RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast.Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):632-46. doi: 10.1002/wrna.80. Epub 2011 Mar 23. Wiley Interdiscip Rev RNA. 2011. PMID: 21823226 Free PMC article. Review.
Cited by
-
A Novel Epigenetic Silencing Pathway Involving the Highly Conserved 5'-3' Exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe.PLoS Genet. 2016 Feb 18;12(2):e1005873. doi: 10.1371/journal.pgen.1005873. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26889830 Free PMC article.
-
WD40 repeats arrange histone tails for spreading of silencing.J Mol Cell Biol. 2010 Apr;2(2):81-3. doi: 10.1093/jmcb/mjp046. Epub 2009 Dec 11. J Mol Cell Biol. 2010. PMID: 20008334 Free PMC article.
-
Set1/COMPASS repels heterochromatin invasion at euchromatic sites by disrupting Suv39/Clr4 activity and nucleosome stability.Genes Dev. 2020 Jan 1;34(1-2):99-117. doi: 10.1101/gad.328468.119. Epub 2019 Dec 5. Genes Dev. 2020. PMID: 31805521 Free PMC article.
-
Stress-sensitive dynamics of miRNAs and Elba1 in Drosophila embryogenesis.Mol Syst Biol. 2023 May 9;19(5):e11148. doi: 10.15252/msb.202211148. Epub 2023 Mar 20. Mol Syst Biol. 2023. PMID: 36938679 Free PMC article.
-
RNA-induced initiation of transcriptional silencing (RITS) complex structure and function.RNA Biol. 2019 Sep;16(9):1133-1146. doi: 10.1080/15476286.2019.1621624. Epub 2019 Jun 18. RNA Biol. 2019. PMID: 31213126 Free PMC article. Review.
References
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. - PubMed
-
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. - PubMed
-
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

