NMDA receptor stimulation induces reversible fission of the neuronal endoplasmic reticulum
- PMID: 19381304
- PMCID: PMC2668765
- DOI: 10.1371/journal.pone.0005250
NMDA receptor stimulation induces reversible fission of the neuronal endoplasmic reticulum
Abstract
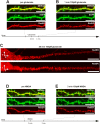
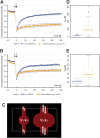
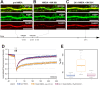
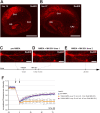
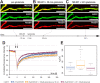
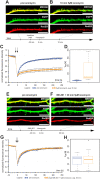
With few exceptions the endoplasmic reticulum (ER) is considered a continuous system of endomembranes within which proteins and ions can move. We have studied dynamic structural changes of the ER in hippocampal neurons in primary culture and organotypic slices. Fluorescence recovery after photobleaching (FRAP) was used to quantify and model ER structural dynamics. Ultrastructure was assessed by electron microscopy. In live cell imaging experiments we found that, under basal conditions, the ER of neuronal soma and dendrites was continuous. The smooth and uninterrupted appearance of the ER changed dramatically after glutamate stimulation. The ER fragmented into isolated vesicles in a rapid fission reaction that occurred prior to overt signs of neuronal damage. ER fission was found to be independent of ER calcium levels. Apart from glutamate, the calcium ionophore ionomycin was able to induce ER fission. The N-methyl, D-aspartate (NMDA) receptor antagonist MK-801 inhibited ER fission induced by glutamate as well as by ionomycin. Fission was not blocked by either ifenprodil or kinase inhibitors. Interestingly, sub-lethal NMDA receptor stimulation caused rapid ER fission followed by fusion. Hence, ER fission is not strictly associated with cellular damage or death. Our results thus demonstrate that neuronal ER structure is dynamically regulated with important consequences for protein mobility and ER luminal calcium tunneling.
Conflict of interest statement
Figures


Similar articles
-
Rapid fragmentation of the endoplasmic reticulum in cortical neurons of the mouse brain in situ following cardiac arrest.J Cereb Blood Flow Metab. 2011 Aug;31(8):1663-7. doi: 10.1038/jcbfm.2011.37. Epub 2011 Apr 6. J Cereb Blood Flow Metab. 2011. PMID: 21468089 Free PMC article.
-
Potassium-induced structural changes of the endoplasmic reticulum in pyramidal neurons in murine organotypic hippocampal slices.J Neurosci Res. 2011 Aug;89(8):1150-9. doi: 10.1002/jnr.22646. Epub 2011 May 2. J Neurosci Res. 2011. PMID: 21538461
-
CaMKII-dependent endoplasmic reticulum fission by whisker stimulation and during cortical spreading depolarization.Brain. 2018 Apr 1;141(4):1049-1062. doi: 10.1093/brain/awy036. Brain. 2018. PMID: 29538620
-
Fission and fusion of the neuronal endoplasmic reticulum.Transl Stroke Res. 2013 Dec;4(6):652-62. doi: 10.1007/s12975-013-0279-9. Epub 2013 Aug 24. Transl Stroke Res. 2013. PMID: 24323419 Review.
-
Mechanisms of Neuronal Protection against Excitotoxicity, Endoplasmic Reticulum Stress, and Mitochondrial Dysfunction in Stroke and Neurodegenerative Diseases.Oxid Med Cell Longev. 2015;2015:964518. doi: 10.1155/2015/964518. Epub 2015 Oct 20. Oxid Med Cell Longev. 2015. PMID: 26576229 Free PMC article. Review.
Cited by
-
Further assembly required: construction and dynamics of the endoplasmic reticulum network.EMBO Rep. 2010 Jul;11(7):515-21. doi: 10.1038/embor.2010.92. Epub 2010 Jun 18. EMBO Rep. 2010. PMID: 20559323 Free PMC article. Review.
-
Endoplasmic Reticulum in Metaplasticity: From Information Processing to Synaptic Proteostasis.Mol Neurobiol. 2022 Sep;59(9):5630-5655. doi: 10.1007/s12035-022-02916-1. Epub 2022 Jun 23. Mol Neurobiol. 2022. PMID: 35739409 Review.
-
Rapid regulation of endoplasmic reticulum dynamics in dendritic spines by NMDA receptor activation.Mol Brain. 2014 Aug 19;7:60. doi: 10.1186/s13041-014-0060-3. Mol Brain. 2014. PMID: 25242397 Free PMC article.
-
Rapid fragmentation of the endoplasmic reticulum in cortical neurons of the mouse brain in situ following cardiac arrest.J Cereb Blood Flow Metab. 2011 Aug;31(8):1663-7. doi: 10.1038/jcbfm.2011.37. Epub 2011 Apr 6. J Cereb Blood Flow Metab. 2011. PMID: 21468089 Free PMC article.
-
Advances in Plant ER Architecture and Dynamics.Plant Physiol. 2018 Jan;176(1):178-186. doi: 10.1104/pp.17.01261. Epub 2017 Oct 6. Plant Physiol. 2018. PMID: 28986423 Free PMC article. Review.
References
-
- Bardo S, Cavazzini MG, Emptage N. The role of the endoplasmic reticulum Ca2+ store in the plasticity of central neurons. Trends Pharmacol Sci. 2006;27:78–84. - PubMed
-
- DeGracia DJ, Montie HL. Cerebral ischemia and the unfolded protein response. J Neurochem. 2004;91:1–8. - PubMed
-
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. - PubMed
-
- Mattson MP. Calcium and neurodegeneration. Aging Cell. 2007;6:337–350. - PubMed
-
- Meldolesi J. Rapidly exchanging Ca2+ stores in neurons: molecular, structural and functional properties. Prog Neurobiol. 2001;65:309–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

