TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma
- PMID: 19380639
- PMCID: PMC2715030
- DOI: 10.1084/jem.20090528
TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma
Abstract
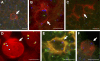
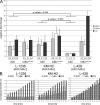
Proliferation and survival of Hodgkin and Reed/Sternberg (HRS) cells, the malignant cells of classical Hodgkin lymphoma (cHL), are dependent on constitutive activation of nuclear factor kappaB (NF-kappaB). NF-kappaB activation through various stimuli is negatively regulated by the zinc finger protein A20. To determine whether A20 contributes to the pathogenesis of cHL, we sequenced TNFAIP3, encoding A20, in HL cell lines and laser-microdissected HRS cells from cHL biopsies. We detected somatic mutations in 16 out of 36 cHLs (44%), including missense mutations in 2 out of 16 Epstein-Barr virus-positive (EBV(+)) cHLs and a missense mutation, nonsense mutations, and frameshift-causing insertions or deletions in 14 out of 20 EBV(-) cHLs. In most mutated cases, both TNFAIP3 alleles were inactivated, including frequent chromosomal deletions of TNFAIP3. Reconstitution of wild-type TNFAIP3 in A20-deficient cHL cell lines revealed a significant decrease in transcripts of selected NF-kappaB target genes and caused cytotoxicity. Extending the mutation analysis to primary mediastinal B cell lymphoma (PMBL), another lymphoma with constitutive NF-kappaB activity, revealed destructive mutations in 5 out of 14 PMBLs (36%). This report identifies TNFAIP3 (A20), a key regulator of NF-kappaB activity, as a novel tumor suppressor gene in cHL and PMBL. The significantly higher frequency of TNFAIP3 mutations in EBV(-) than EBV(+) cHL suggests complementing functions of TNFAIP3 inactivation and EBV infection in cHL pathogenesis.
Figures

Similar articles
-
A20 (TNFAIP3) deletion in Epstein-Barr virus-associated lymphoproliferative disorders/lymphomas.PLoS One. 2013;8(2):e56741. doi: 10.1371/journal.pone.0056741. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418597 Free PMC article.
-
Mutation analysis of tumor necrosis factor alpha-induced protein 3 gene in Hodgkin lymphoma.Pathol Res Pract. 2017 Mar;213(3):256-260. doi: 10.1016/j.prp.2016.11.001. Epub 2016 Nov 26. Pathol Res Pract. 2017. PMID: 28189285
-
Rare occurrence of biallelic CYLD gene mutations in classical Hodgkin lymphoma.Genes Chromosomes Cancer. 2010 Sep;49(9):803-9. doi: 10.1002/gcc.20789. Genes Chromosomes Cancer. 2010. PMID: 20607853
-
Molecular biology of Hodgkin lymphoma.Hematology Am Soc Hematol Educ Program. 2009:491-6. doi: 10.1182/asheducation-2009.1.491. Hematology Am Soc Hematol Educ Program. 2009. PMID: 20008234 Review.
-
A20 takes on tumors: tumor suppression by an ubiquitin-editing enzyme.J Exp Med. 2009 May 11;206(5):977-80. doi: 10.1084/jem.20090765. Epub 2009 Apr 20. J Exp Med. 2009. PMID: 19380636 Free PMC article. Review.
Cited by
-
A20 (TNFAIP3) deletion in Epstein-Barr virus-associated lymphoproliferative disorders/lymphomas.PLoS One. 2013;8(2):e56741. doi: 10.1371/journal.pone.0056741. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418597 Free PMC article.
-
Somatic mutations in autoinflammatory and autoimmune disease.Nat Rev Rheumatol. 2024 Nov;20(11):683-698. doi: 10.1038/s41584-024-01168-8. Epub 2024 Oct 11. Nat Rev Rheumatol. 2024. PMID: 39394526 Review.
-
A20 promotes melanoma progression via the activation of Akt pathway.Cell Death Dis. 2020 Sep 23;11(9):794. doi: 10.1038/s41419-020-03001-y. Cell Death Dis. 2020. PMID: 32968045 Free PMC article.
-
Expression of A20 is reduced in pancreatic cancer tissues.J Mol Histol. 2012 Jun;43(3):319-25. doi: 10.1007/s10735-012-9402-6. Epub 2012 Mar 30. J Mol Histol. 2012. PMID: 22461193
-
Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets.Cancer Biol Ther. 2010 Oct 15;10(8):737-47. doi: 10.4161/cbt.10.8.13417. Epub 2010 Oct 15. Cancer Biol Ther. 2010. PMID: 20930542 Free PMC article. Review.
References
-
- Bargou R.C., Emmerich F., Krappmann D., Bommert K., Mapara M.Y., Arnold W., Royer H.D., Grinstein E., Greiner A., Scheidereit C., Dorken B. 1997. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells.J. Clin. Invest. 100:2961–2969 - PMC - PubMed
-
- Hinz M., Lemke P., Anagnostopoulos I., Hacker C., Krappmann D., Mathas S., Dorken B., Zenke M., Stein H., Scheidereit C. 2002. Nuclear factor κB–dependent gene expression profiling of Hodgkin’s disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity.J. Exp. Med. 196:605–617 - PMC - PubMed
-
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling.Cell. 132:344–362 - PubMed
-
- Wu C.J., Conze D.B., Li T., Srinivasula S.M., Ashwell J.D. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation.Nat. Cell Biol. 8:398–406 - PubMed
-
- Ea C.K., Deng L., Xia Z.P., Pineda G., Chen Z.J. 2006. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO.Mol. Cell. 22:245–257 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

