Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription
- PMID: 19359585
- PMCID: PMC2785900
- DOI: 10.1126/science.1164860
Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription
Abstract
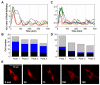
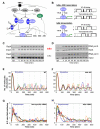
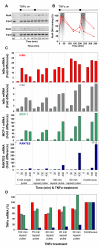
The nuclear factor kappaB (NF-kappaB) transcription factor regulates cellular stress responses and the immune response to infection. NF-kappaB activation results in oscillations in nuclear NF-kappaB abundance. To define the function of these oscillations, we treated cells with repeated short pulses of tumor necrosis factor-alpha at various intervals to mimic pulsatile inflammatory signals. At all pulse intervals that were analyzed, we observed synchronous cycles of NF-kappaB nuclear translocation. Lower frequency stimulations gave repeated full-amplitude translocations, whereas higher frequency pulses gave reduced translocation, indicating a failure to reset. Deterministic and stochastic mathematical models predicted how negative feedback loops regulate both the resetting of the system and cellular heterogeneity. Altering the stimulation intervals gave different patterns of NF-kappaB-dependent gene expression, which supports the idea that oscillation frequency has a functional role.
Figures

Similar articles
-
Oscillations in NF-kappaB signaling control the dynamics of gene expression.Science. 2004 Oct 22;306(5696):704-8. doi: 10.1126/science.1099962. Science. 2004. PMID: 15499023
-
Spatial and temporal information coding and noise in the NF-κB system.Biochem Soc Trans. 2010 Oct;38(5):1247-50. doi: 10.1042/BST0381247. Biochem Soc Trans. 2010. PMID: 20863293
-
Physiological levels of TNFalpha stimulation induce stochastic dynamics of NF-kappaB responses in single living cells.J Cell Sci. 2010 Aug 15;123(Pt 16):2834-43. doi: 10.1242/jcs.069641. Epub 2010 Jul 27. J Cell Sci. 2010. PMID: 20663918 Free PMC article.
-
Duration of nuclear NF-kappaB action regulated by reversible acetylation.Science. 2001 Aug 31;293(5535):1653-7. doi: 10.1126/science.1062374. Science. 2001. PMID: 11533489
-
Comment on "Oscillations in NF-kappaB signaling control the dynamics of gene expression".Science. 2005 Apr 1;308(5718):52; author reply 52. doi: 10.1126/science.1108198. Science. 2005. PMID: 15802586 Free PMC article. No abstract available.
Cited by
-
Automated analysis of NF-κB nuclear translocation kinetics in high-throughput screening.PLoS One. 2012;7(12):e52337. doi: 10.1371/journal.pone.0052337. Epub 2012 Dec 27. PLoS One. 2012. PMID: 23300644 Free PMC article.
-
Tension and robustness in multitasking cellular networks.PLoS Comput Biol. 2012;8(4):e1002491. doi: 10.1371/journal.pcbi.1002491. Epub 2012 Apr 26. PLoS Comput Biol. 2012. PMID: 22577355 Free PMC article.
-
Efficacy of Artemisia annua L. extract for recovery of acute liver failure.Food Sci Nutr. 2020 Jun 5;8(7):3738-3749. doi: 10.1002/fsn3.1662. eCollection 2020 Jul. Food Sci Nutr. 2020. PMID: 32724636 Free PMC article.
-
A systematic survey of the response of a model NF-κB signalling pathway to TNFα stimulation.J Theor Biol. 2012 Mar 21;297(2-12):137-47. doi: 10.1016/j.jtbi.2011.12.014. Epub 2011 Dec 23. J Theor Biol. 2012. PMID: 22202812 Free PMC article.
-
Immunological synapse: a multi-protein signalling cellular apparatus for controlling gene expression.Immunology. 2010 Mar;129(3):322-8. doi: 10.1111/j.1365-2567.2009.03241.x. Immunology. 2010. PMID: 20409153 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/E012965/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBC0071581/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C520471/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D010748/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E004210/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C007158/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C008219/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0500346(73596)/MRC_/Medical Research Council/United Kingdom
- BBF0059381/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBC0082191/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0500346/MRC_/Medical Research Council/United Kingdom
- BBD0107481/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBC5204711/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F005938/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

