Structural and functional similarities between a ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)-like protein from Bacillus subtilis and photosynthetic RuBisCO
- PMID: 19279009
- PMCID: PMC2676057
- DOI: 10.1074/jbc.M807095200
Structural and functional similarities between a ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)-like protein from Bacillus subtilis and photosynthetic RuBisCO
Abstract
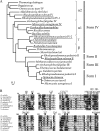
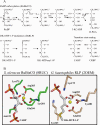
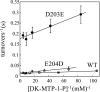
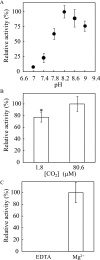
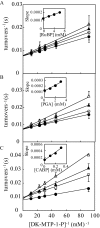
The sequences classified as genes for various ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (RuBisCO)-like proteins (RLPs) are widely distributed among bacteria, archaea, and eukaryota. In the phylogenic tree constructed with these sequences, RuBisCOs and RLPs are grouped into four separate clades, forms I-IV. In RuBisCO enzymes encoded by form I, II, and III sequences, 19 conserved amino acid residues are essential for CO(2) fixation; however, 1-11 of these 19 residues are substituted with other amino acids in form IV RLPs. Among form IV RLPs, the only enzymatic activity detected to date is a 2,3-diketo-5-methylthiopentyl 1-phosphate (DK-MTP-1-P) enolase reaction catalyzed by Bacillus subtilis, Microcystis aeruginosa, and Geobacillus kaustophilus form IV RLPs. RLPs from Rhodospirillum rubrum, Rhodopseudomonas palustris, Chlorobium tepidum, and Bordetella bronchiseptica were inactive in the enolase reaction. DK-MTP-1-P enolase activity of B. subtilis RLP required Mg(2+) for catalysis and, like RuBisCO, was stimulated by CO(2). Four residues that are essential for the enolization reaction of RuBisCO, Lys(175), Lys(201), Asp(203), and Glu(204), were conserved in RLPs and were essential for DK-MTP-1-P enolase catalysis. Lys(123), the residue conserved in DK-MTP-1-P enolases, was also essential for B. subtilis RLP enolase activity. Similarities between the active site structures of RuBisCO and B. subtilis RLP were examined by analyzing the effects of structural analogs of RuBP on DK-MTP-1-P enolase activity. A transition state analog for the RuBP carboxylation of RuBisCO was a competitive inhibitor in the DK-MTP-1-P enolase reaction with a K(i) value of 103 mum. RuBP and d-phosphoglyceric acid, the substrate and product, respectively, of RuBisCO, were weaker competitive inhibitors. These results suggest that the amino acid residues utilized in the B. subtilis RLP enolase reaction are the same as those utilized in the RuBisCO RuBP enolization reaction.
Figures
Similar articles
-
RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO.J Exp Bot. 2008;59(7):1543-54. doi: 10.1093/jxb/ern104. Epub 2008 Apr 9. J Exp Bot. 2008. PMID: 18403380
-
Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis.Acta Crystallogr D Biol Crystallogr. 2009 Sep;65(Pt 9):942-51. doi: 10.1107/S0907444909022422. Epub 2009 Aug 14. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19690372
-
Mechanistic diversity in the RuBisCO superfamily: the "enolase" in the methionine salvage pathway in Geobacillus kaustophilus.Biochemistry. 2007 Apr 3;46(13):4077-89. doi: 10.1021/bi7000483. Epub 2007 Mar 13. Biochemistry. 2007. PMID: 17352497
-
Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism?Res Microbiol. 2005 Jun-Jul;156(5-6):611-8. doi: 10.1016/j.resmic.2005.01.014. Epub 2005 Feb 26. Res Microbiol. 2005. PMID: 15950120 Review.
-
Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs.Microbiol Mol Biol Rev. 2007 Dec;71(4):576-99. doi: 10.1128/MMBR.00015-07. Microbiol Mol Biol Rev. 2007. PMID: 18063718 Free PMC article. Review.
Cited by
-
Metabolic Regulation as a Consequence of Anaerobic 5-Methylthioadenosine Recycling in Rhodospirillum rubrum.mBio. 2016 Jul 12;7(4):e00855-16. doi: 10.1128/mBio.00855-16. mBio. 2016. PMID: 27406564 Free PMC article.
-
MtnBD is a multifunctional fusion enzyme in the methionine salvage pathway of Tetrahymena thermophila.PLoS One. 2013 Jul 1;8(7):e67385. doi: 10.1371/journal.pone.0067385. Print 2013. PLoS One. 2013. PMID: 23840871 Free PMC article.
-
Phosphates as Energy Sources to Expand Metabolic Networks.Life (Basel). 2019 May 22;9(2):43. doi: 10.3390/life9020043. Life (Basel). 2019. PMID: 31121973 Free PMC article.
-
A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic.J Biol Chem. 2017 Apr 21;292(16):6838-6850. doi: 10.1074/jbc.M116.767145. Epub 2017 Jan 30. J Biol Chem. 2017. PMID: 28154188 Free PMC article.
-
Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco.Plant Physiol. 2011 Jan;155(1):27-35. doi: 10.1104/pp.110.164814. Epub 2010 Oct 25. Plant Physiol. 2011. PMID: 20974895 Free PMC article. No abstract available.
References
-
- Miziorko, H. M., and Lorimer, G. H. (1983) Annu. Rev. Biochem. 52 507-535 - PubMed
-
- Andrews, T. J., and Lorimer, G. H. (1987) in The Biochemistry of Plants (Hatch, M. D., and Boardman, N. K., eds) Vol. 10, pp. 131-218, Academic Press, Inc., New York
-
- Hartman, F. C., and Harpel, M. R. (1994) Annu. Rev. Biochem. 63 197-234 - PubMed
-
- Portis, A. R., Jr., and Parry, M. A. (2007) Photosynth. Res. 94 121-143 - PubMed
-
- Roy, H., and Andrews, T. J. (2000) in Photosynthesis: Physiology and Metabolism (Leegood, R. C., Sharkey, T. D., and von Caemmerer, S., eds) pp. 53-83, Kluwer Academic, Dordrecht, The Netherlands
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

