The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans
- PMID: 19273594
- PMCID: PMC2682033
- DOI: 10.1128/MCB.01811-08
The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans
Abstract
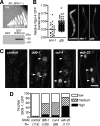
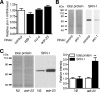
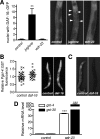
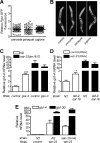
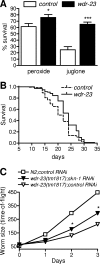
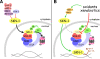
The transcription factor SKN-1 protects Caenorhabditis elegans from stress and promotes longevity. SKN-1 is regulated by diverse signals that control metabolism, development, and stress responses, but the mechanisms of regulation and signal integration are unknown. We screened the C. elegans genome for regulators of cytoprotective gene expression and identified a new SKN-1 regulatory pathway. SKN-1 protein levels, nuclear accumulation, and activity are repressed by the WD40 repeat protein WDR-23, which interacts with the CUL-4/DDB-1 ubiquitin ligase to presumably target the transcription factor for proteasomal degradation. WDR-23 regulates SKN-1 target genes downstream from p38 mitogen-activated protein kinase, glycogen synthase kinase 3, and insulin-like receptor pathways, suggesting that phosphorylation of SKN-1 may function to modify its interaction with WDR-23 and/or CUL-4/DDB-1. These findings define the mechanism of SKN-1 accumulation in the cell nucleus and provide a new mechanistic framework for understanding how phosphorylation signals are integrated to regulate stress resistance and longevity.
Figures
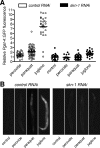
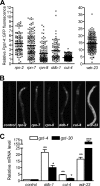

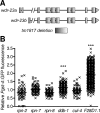
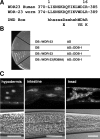
Similar articles
-
Direct interaction between the WD40 repeat protein WDR-23 and SKN-1/Nrf inhibits binding to target DNA.Mol Cell Biol. 2014 Aug;34(16):3156-67. doi: 10.1128/MCB.00114-14. Epub 2014 Jun 9. Mol Cell Biol. 2014. Retraction in: Mol Cell Biol. 2015 Sep;35(18):3255. doi: 10.1128/MCB.00658-15. PMID: 24912676 Free PMC article. Retracted.
-
Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleiotrophy, aging, glutathione, and interactions with other longevity pathways.Mech Ageing Dev. 2015 Jul;149:88-98. doi: 10.1016/j.mad.2015.06.001. Epub 2015 Jun 6. Mech Ageing Dev. 2015. PMID: 26056713
-
The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans.PLoS Genet. 2013 Mar;9(3):e1003354. doi: 10.1371/journal.pgen.1003354. Epub 2013 Mar 21. PLoS Genet. 2013. PMID: 23555279 Free PMC article.
-
SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans.Free Radic Biol Med. 2015 Nov;88(Pt B):290-301. doi: 10.1016/j.freeradbiomed.2015.06.008. Epub 2015 Aug 5. Free Radic Biol Med. 2015. PMID: 26232625 Free PMC article. Review.
-
Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance.Drug Metab Rev. 2012 Aug;44(3):209-23. doi: 10.3109/03602532.2012.684799. Epub 2012 Jun 4. Drug Metab Rev. 2012. PMID: 22656429 Free PMC article. Review.
Cited by
-
Mechanisms and functions of Nrf2 signaling in Drosophila.Free Radic Biol Med. 2015 Nov;88(Pt B):302-313. doi: 10.1016/j.freeradbiomed.2015.06.020. Epub 2015 Jun 25. Free Radic Biol Med. 2015. PMID: 26117322 Free PMC article. Review.
-
Increased expression of metabolism and lysosome-associated genes in a C. elegans dpy-7 cuticle furrow mutant.MicroPubl Biol. 2024 Jul 31;2024:10.17912/micropub.biology.001241. doi: 10.17912/micropub.biology.001241. eCollection 2024. MicroPubl Biol. 2024. PMID: 39144098 Free PMC article.
-
SKN-1 Is a Negative Regulator of DAF-16 and Somatic Stress Resistance in Caenorhabditis elegans.G3 (Bethesda). 2020 May 4;10(5):1707-1712. doi: 10.1534/g3.120.401203. G3 (Bethesda). 2020. PMID: 32161088 Free PMC article.
-
Regulation of synaptic nlg-1/neuroligin abundance by the skn-1/Nrf stress response pathway protects against oxidative stress.PLoS Genet. 2014 Jan;10(1):e1004100. doi: 10.1371/journal.pgen.1004100. Epub 2014 Jan 16. PLoS Genet. 2014. PMID: 24453991 Free PMC article.
-
The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans.Aging Cell. 2014 Feb;13(1):70-9. doi: 10.1111/acel.12154. Epub 2013 Sep 18. Aging Cell. 2014. PMID: 23957350 Free PMC article.
References
-
- Angers, S., T. Li, X. Yi, M. J. MacCoss, R. T. Moon, and N. Zheng. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443590-593. - PubMed
-
- Bernhardt, A., E. Lechner, P. Hano, V. Schade, M. Dieterle, M. Anders, M. J. Dubin, G. Benvenuto, C. Bowler, P. Genschik, and H. Hellmann. 2006. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47591-603. - PubMed
-
- Bishop, N. A., and L. Guarente. 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447545-549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007628/GM/NIGMS NIH HHS/United States
- R01 GM042056/GM/NIGMS NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 DK061168/DK/NIDDK NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- F32 GM077904/GM/NIGMS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- R01 DK61168/DK/NIDDK NIH HHS/United States
- HD15052/HD/NICHD NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
- R01 GM42056/GM/NIGMS NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- GM077904/GM/NIGMS NIH HHS/United States
- DK58404/DK/NIDDK NIH HHS/United States
- EY08126/EY/NEI NIH HHS/United States
- T32 GM07628/GM/NIGMS NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- R37 GM042056/GM/NIGMS NIH HHS/United States
- DK59637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
