Allostery in the LacI/GalR family: variations on a theme
- PMID: 19269243
- PMCID: PMC2688824
- DOI: 10.1016/j.mib.2009.01.009
Allostery in the LacI/GalR family: variations on a theme
Abstract
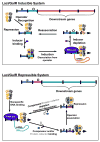
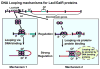
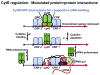
The lactose repressor protein (LacI) was among the very first genetic regulatory proteins discovered, and more than 1000 members of the bacterial LacI/GalR family are now identified. LacI has been the prototype for understanding how transcription is controlled using small metabolites to modulate protein association with specific DNA sites. This understanding has been greatly expanded by the study of other LacI/GalR homologues. A general picture emerges in which the conserved fold provides a scaffold for multiple types of interactions - including oligomerization, small molecule binding, and protein-protein binding - that in turn influence target DNA binding and thereby regulate mRNA production. Although many different functions have evolved from this basic scaffold, each homologue retains functional flexibility: For the same protein, different small molecules can have disparate impact on DNA binding and hence transcriptional outcome. In turn, binding to alternative DNA sequences may impact the degree of allosteric response. Thus, this family exhibits a symphony of variations by which transcriptional control is achieved.
Figures
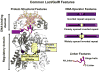
Comment in
-
Small molecule signaling.Curr Opin Microbiol. 2009 Apr;12(2):125-8. doi: 10.1016/j.mib.2009.02.002. Epub 2009 Mar 9. Curr Opin Microbiol. 2009. PMID: 19278897 No abstract available.
Similar articles
-
Subdividing repressor function: DNA binding affinity, selectivity, and allostery can be altered by amino acid substitution of nonconserved residues in a LacI/GalR homologue.Biochemistry. 2008 Aug 5;47(31):8058-69. doi: 10.1021/bi800443k. Epub 2008 Jul 11. Biochemistry. 2008. PMID: 18616293 Free PMC article.
-
Novel insights from hybrid LacI/GalR proteins: family-wide functional attributes and biologically significant variation in transcription repression.Nucleic Acids Res. 2012 Nov;40(21):11139-54. doi: 10.1093/nar/gks806. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965134 Free PMC article.
-
Flexibility in the inducer binding region is crucial for allostery in the Escherichia coli lactose repressor.Biochemistry. 2009 Jun 9;48(22):4988-98. doi: 10.1021/bi9002343. Biochemistry. 2009. PMID: 19368358 Free PMC article.
-
Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors.FEBS Lett. 1995 Dec 18;377(2):98-102. doi: 10.1016/0014-5793(95)01344-x. FEBS Lett. 1995. PMID: 8543068 Review.
-
Flexibility and Disorder in Gene Regulation: LacI/GalR and Hox Proteins.J Biol Chem. 2015 Oct 9;290(41):24669-77. doi: 10.1074/jbc.R115.685032. Epub 2015 Sep 4. J Biol Chem. 2015. PMID: 26342073 Free PMC article. Review.
Cited by
-
Comparative genomics and transcriptomics analyses provide insights into the high yield and regulatory mechanism of Norvancomycin biosynthesis in Amycolatopsis orientalis NCPC 2-48.Microb Cell Fact. 2021 Feb 2;20(1):28. doi: 10.1186/s12934-021-01521-6. Microb Cell Fact. 2021. PMID: 33531006 Free PMC article.
-
Engineering Allostery into Proteins.Adv Exp Med Biol. 2019;1163:359-384. doi: 10.1007/978-981-13-8719-7_15. Adv Exp Med Biol. 2019. PMID: 31707711 Free PMC article. Review.
-
Accelerating Genetic Sensor Development, Scale-up, and Deployment Using Synthetic Biology.Biodes Res. 2024 Jun 25;6:0037. doi: 10.34133/bdr.0037. eCollection 2024. Biodes Res. 2024. PMID: 38919711 Free PMC article.
-
Reconciling kinetic and thermodynamic models of bacterial transcription.PLoS Comput Biol. 2021 Jan 19;17(1):e1008572. doi: 10.1371/journal.pcbi.1008572. eCollection 2021 Jan. PLoS Comput Biol. 2021. PMID: 33465069 Free PMC article.
-
Insights into the regulatory mechanisms and application prospects of the transcription factor Cra.Appl Environ Microbiol. 2024 Nov 20;90(11):e0122824. doi: 10.1128/aem.01228-24. Epub 2024 Nov 4. Appl Environ Microbiol. 2024. PMID: 39494897 Review.
References
-
- Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. - PubMed
-
- Colmer JA, Hamood AN. Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR. Mol Gen Genet. 1998;258:250–259. - PubMed
-
- Meng LM, Nygaard P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol Microbiol. 1990;4:2187–2192. - PubMed
-
- Shelburne SA, III, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2008;105:1698–1703. In group A Streptococcus, transcription of several virulence factors is related to carbohydrate utilization. Bacterial variants with a deleted CcpA gene show decreased virulence in a mouse model of invasive infection and changed expression of virulence genes on a nutrient-limited growth media. Further, purified CcpA bound directly to the promoter region of the virulence gene for steptolysin S. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

