Requirements for NuMA in maintenance and establishment of mammalian spindle poles
- PMID: 19255246
- PMCID: PMC2686415
- DOI: 10.1083/jcb.200810091
Requirements for NuMA in maintenance and establishment of mammalian spindle poles
Abstract
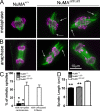
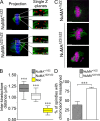
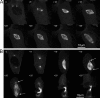
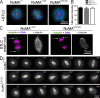
Microtubules of the mitotic spindle in mammalian somatic cells are focused at spindle poles, a process thought to include direct capture by astral microtubules of kinetochores and/or noncentrosomally nucleated microtubule bundles. By construction and analysis of a conditional loss of mitotic function allele of the nuclear mitotic apparatus (NuMA) protein in mice and cultured primary cells, we demonstrate that NuMA is an essential mitotic component with distinct contributions to the establishment and maintenance of focused spindle poles. When mitotic NuMA function is disrupted, centrosomes provide initial focusing activity, but continued centrosome attachment to spindle fibers under tension is defective, and the maintenance of focused kinetochore fibers at spindle poles throughout mitosis is prevented. Without centrosomes and NuMA, initial establishment of spindle microtubule focusing completely fails. Thus, NuMA is a defining feature of the mammalian spindle pole and functions as an essential tether linking bulk microtubules of the spindle to centrosomes.
Figures


Similar articles
-
NuMA deficiency causes micronuclei via checkpoint-insensitive k-fiber minus-end detachment from mitotic spindle poles.Curr Biol. 2023 Feb 6;33(3):572-580.e2. doi: 10.1016/j.cub.2022.12.017. Epub 2023 Jan 9. Curr Biol. 2023. PMID: 36626904
-
Chromosome movement in mitosis requires microtubule anchorage at spindle poles.J Cell Biol. 2001 Feb 5;152(3):425-34. doi: 10.1083/jcb.152.3.425. J Cell Biol. 2001. PMID: 11157972 Free PMC article.
-
Nucleoporin Nup188 is required for chromosome alignment in mitosis.Cancer Sci. 2013 Jul;104(7):871-9. doi: 10.1111/cas.12159. Epub 2013 May 2. Cancer Sci. 2013. PMID: 23551833 Free PMC article.
-
Cooperative mechanisms of mitotic spindle formation.J Cell Sci. 2007 May 15;120(Pt 10):1717-22. doi: 10.1242/jcs.03442. J Cell Sci. 2007. PMID: 17502482 Review.
-
Cell and molecular biology of spindle poles and NuMA.Int Rev Cytol. 2004;238:1-57. doi: 10.1016/S0074-7696(04)38001-0. Int Rev Cytol. 2004. PMID: 15364196 Review.
Cited by
-
WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression.J Cell Sci. 2012 Nov 1;125(Pt 21):5096-109. doi: 10.1242/jcs.107326. Epub 2012 Aug 16. J Cell Sci. 2012. PMID: 22899712 Free PMC article.
-
Control of Intestinal Cell Fate by Dynamic Mitotic Spindle Repositioning Influences Epithelial Homeostasis and Longevity.Cell Rep. 2019 Sep 10;28(11):2807-2823.e5. doi: 10.1016/j.celrep.2019.08.014. Cell Rep. 2019. PMID: 31509744 Free PMC article.
-
Mitotic spindle proteomics in Chinese hamster ovary cells.PLoS One. 2011;6(5):e20489. doi: 10.1371/journal.pone.0020489. Epub 2011 May 27. PLoS One. 2011. PMID: 21647379 Free PMC article.
-
BetaHPV E6 and E7 colocalize with NuMa in dividing keratinocytes.Virus Genes. 2019 Oct;55(5):600-609. doi: 10.1007/s11262-019-01685-9. Epub 2019 Jul 9. Virus Genes. 2019. PMID: 31290065
-
Signal-induced PARP1-Erk synergism mediates IEG expression.Signal Transduct Target Ther. 2019 Apr 12;4:8. doi: 10.1038/s41392-019-0042-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 30993015 Free PMC article. Review.
References
-
- Chatzimeletiou K., Morrison E.E., Prapas N., Prapas Y., Handyside A.H. 2008. The centrosome and early embryogenesis: clinical insights.Reprod. Biomed. Online. 16:485–491 - PubMed
-
- Cheeseman I.M., Desai A. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans.Sci. STKE. - PubMed
-
- Compton D.A. 1998. Focusing on spindle poles.J. Cell Sci. 111(Pt 11)1477–1481 - PubMed
-
- Dasso M. 2001. Running on Ran: nuclear transport and the mitotic spindle.Cell. 104:321–324 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

