Covalently bound substrate at the regulatory site of yeast pyruvate decarboxylases triggers allosteric enzyme activation
- PMID: 19246454
- PMCID: PMC2673282
- DOI: 10.1074/jbc.M806228200
Covalently bound substrate at the regulatory site of yeast pyruvate decarboxylases triggers allosteric enzyme activation
Abstract
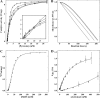
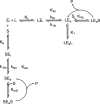
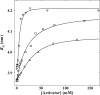
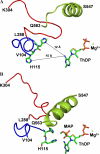
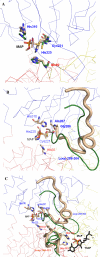
The mechanism by which the enzyme pyruvate decarboxylase from two yeast species is activated allosterically has been elucidated. A total of seven three-dimensional structures of the enzyme, of enzyme variants, or of enzyme complexes from two yeast species, three of them reported here for the first time, provide detailed atomic resolution snapshots along the activation coordinate. The prime event is the covalent binding of the substrate pyruvate to the side chain of cysteine 221, thus forming a thiohemiketal. This reaction causes the shift of a neighboring amino acid, which eventually leads to the rigidification of two otherwise flexible loops, one of which provides two histidine residues necessary to complete the enzymatically competent active site architecture. The structural data are complemented and supported by kinetic investigations and binding studies, providing a consistent picture of the structural changes occurring upon enzyme activation.
Figures
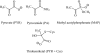


Similar articles
-
The crystal structure of pyruvate decarboxylase from Kluyveromyces lactis. Implications for the substrate activation mechanism of this enzyme.FEBS J. 2006 Sep;273(18):4199-209. doi: 10.1111/j.1742-4658.2006.05415.x. FEBS J. 2006. PMID: 16939618
-
Pyruvate decarboxylase from Kluyveromyces lactis. An enzyme with an extraordinary substrate activation behaviour.Eur J Biochem. 2002 Jul;269(13):3256-63. doi: 10.1046/j.1432-1033.2002.03006.x. Eur J Biochem. 2002. PMID: 12084066
-
Significance of Individual Residues at the Regulatory Site of Yeast Pyruvate Decarboxylase for Allosteric Substrate Activation.Biochemistry. 2017 Mar 7;56(9):1285-1298. doi: 10.1021/acs.biochem.6b01158. Epub 2017 Feb 23. Biochemistry. 2017. PMID: 28170226
-
Regulation of thiamin diphosphate-dependent 2-oxo acid decarboxylases by substrate and thiamin diphosphate.Mg(II) - evidence for tertiary and quaternary interactions.Biochim Biophys Acta. 1998 Jun 29;1385(2):287-306. doi: 10.1016/s0167-4838(98)00075-2. Biochim Biophys Acta. 1998. PMID: 9655921 Review.
-
The structure and the mechanism of action of pyruvate carboxylase.Int J Biochem Cell Biol. 1995 Mar;27(3):231-49. doi: 10.1016/1357-2725(94)00087-r. Int J Biochem Cell Biol. 1995. PMID: 7780827 Review.
Cited by
-
Biochemical Characterization and Structural Insight into Interaction and Conformation Mechanisms of Serratia marcescens Lysine Decarboxylase (SmcadA).Molecules. 2021 Jan 29;26(3):697. doi: 10.3390/molecules26030697. Molecules. 2021. PMID: 33572696 Free PMC article.
-
Enzymes for Efficient CO2 Conversion.Protein J. 2021 Aug;40(4):489-503. doi: 10.1007/s10930-021-10007-8. Epub 2021 Jun 7. Protein J. 2021. PMID: 34100161 Review.
-
Assignment of PolyProline II conformation and analysis of sequence--structure relationship.PLoS One. 2011 Mar 31;6(3):e18401. doi: 10.1371/journal.pone.0018401. PLoS One. 2011. PMID: 21483785 Free PMC article.
-
Perturbation of the monomer-monomer interfaces of the benzoylformate decarboxylase tetramer.Biochemistry. 2014 Jul 15;53(27):4358-67. doi: 10.1021/bi500081r. Epub 2014 Jul 3. Biochemistry. 2014. PMID: 24956165 Free PMC article.
-
Allosteric inhibition of Staphylococcus aureus MenD by 1,4-dihydroxy naphthoic acid: a feedback inhibition mechanism of the menaquinone biosynthesis pathway.Philos Trans R Soc Lond B Biol Sci. 2023 Feb 27;378(1871):20220035. doi: 10.1098/rstb.2022.0035. Epub 2023 Jan 11. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 36633276 Free PMC article.
References
-
- Baburina, I., Gao, Y., Hu, Z., Jordan, F., Hohmann, S., and Furey, W. (1994) Biochemistry 33 5630-5635 - PubMed
-
- Killenberg-Jabs, M., König, S., Hohmann, S., and Hübner, G. (1996) Biol. Chem. Hoppe-Seyler 377 313-317 - PubMed
-
- Killenberg-Jabs, M., König, S., Eberhardt, I., Hohmann, S., and Hübner, G. (1997) Biochemistry 36 1900-1905 - PubMed
-
- Baburina, I., Li, H., Bennion, B., Furey, W., and Jordan, F. (1998) Biochemistry 37 1235-1244 - PubMed
-
- Li, H., Furey, W., and Jordan, F. (1999) Biochemistry 38 9992-10003 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

