The Accessory SecA2 System of Mycobacteria Requires ATP Binding and the Canonical SecA1
- PMID: 19240020
- PMCID: PMC2665116
- DOI: 10.1074/jbc.M900325200
The Accessory SecA2 System of Mycobacteria Requires ATP Binding and the Canonical SecA1
Abstract
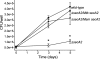
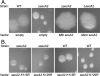
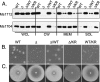
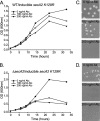
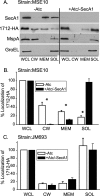
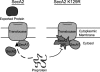
In bacteria, the majority of exported proteins are transported by the general Sec pathway from their site of synthesis in the cytoplasm across the cytoplasmic membrane. The essential SecA ATPase powers this Sec-mediated export. Mycobacteria possess two nonredundant SecA homologs: SecA1 and SecA2. In pathogenic Mycobacterium tuberculosis and the nonpathogenic model mycobacterium Mycobacterium smegmatis, SecA1 is essential for protein export and is the "housekeeping" SecA, whereas SecA2 is an accessory SecA that exports a specific subset of proteins. In M. tuberculosis the accessory SecA2 pathway plays a role in virulence. In this study, we uncovered basic properties of the mycobacterial SecA2 protein and its pathway for exporting select proteins. By constructing secA2 mutant alleles that encode proteins defective in ATP binding, we showed that ATP binding is required for SecA2 function. SecA2 mutant proteins unable to bind ATP were nonfunctional and dominant negative. By evaluating the subcellular distribution of each SecA, SecA1 was shown to be equally divided between cytosolic and cell envelope fractions, whereas SecA2 was predominantly localized to the cytosol. Finally, we showed that the canonical SecA1 has a role in the process of SecA2-dependent export. The accessory SecA2 export system is important to the physiology and virulence of mycobacteria. These studies help establish the mechanism of this new type of specialized protein export pathway.
Figures
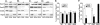
Similar articles
-
Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.J Bacteriol. 2015 Dec 14;198(4):720-30. doi: 10.1128/JB.00696-15. J Bacteriol. 2015. PMID: 26668263 Free PMC article.
-
ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages.J Bacteriol. 2008 Jul;190(14):4880-7. doi: 10.1128/JB.00412-08. Epub 2008 May 16. J Bacteriol. 2008. PMID: 18487341 Free PMC article.
-
Two nonredundant SecA homologues function in mycobacteria.J Bacteriol. 2001 Dec;183(24):6979-90. doi: 10.1128/JB.183.24.6979-6990.2001. J Bacteriol. 2001. PMID: 11717254 Free PMC article.
-
Emerging themes in SecA2-mediated protein export.Nat Rev Microbiol. 2012 Nov;10(11):779-89. doi: 10.1038/nrmicro2874. Epub 2012 Sep 24. Nat Rev Microbiol. 2012. PMID: 23000954 Free PMC article. Review.
-
The Sec Pathways and Exportomes of Mycobacterium tuberculosis.Microbiol Spectr. 2017 Apr;5(2). doi: 10.1128/microbiolspec.TBTB2-0013-2016. Microbiol Spectr. 2017. PMID: 28387178 Review.
Cited by
-
Mycobacterium tuberculosis SatS is a chaperone for the SecA2 protein export pathway.Elife. 2019 Jan 3;8:e40063. doi: 10.7554/eLife.40063. Elife. 2019. PMID: 30604681 Free PMC article.
-
Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain.J Bacteriol. 2013 Feb;195(4):672-81. doi: 10.1128/JB.02032-12. Epub 2012 Nov 30. J Bacteriol. 2013. PMID: 23204463 Free PMC article.
-
Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.J Bacteriol. 2015 Dec 14;198(4):720-30. doi: 10.1128/JB.00696-15. J Bacteriol. 2015. PMID: 26668263 Free PMC article.
-
Mycobacterium tuberculosis SecA2-dependent activation of host Rig-I/MAVs signaling is not conserved in Mycobacterium marinum.PLoS One. 2024 Feb 23;19(2):e0281564. doi: 10.1371/journal.pone.0281564. eCollection 2024. PLoS One. 2024. PMID: 38394154 Free PMC article.
-
Regulated Expression Systems for Mycobacteria and Their Applications.Microbiol Spectr. 2014;2(1):03. doi: 10.1128/microbiolspec.MGM2-0018-2013. Microbiol Spectr. 2014. PMID: 25485177 Free PMC article.
References
-
- Abdallah, A. M., Gey van Pittius, N. C., Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M., Appelmelk, B. J., and Bitter, W. (2007) Nat. Rev. Microbiol. 5 883–891 - PubMed
-
- Gerlach, R. G., and Hensel, M. (2007) Int. J. Med. Microbiol. 297 401–415 - PubMed
-
- Braunstein, M., Espinosa, B., Chan, J., Belisle, J. T., and Jacobs, W. R. J. (2003) Mol. Microbiol. 48 453–464 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

