A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix
- PMID: 19229133
- PMCID: PMC2751576
- DOI: 10.4161/cc.8.6.7941
A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix
Abstract
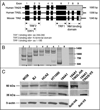
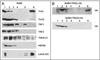
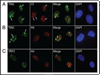
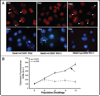
Telomeres are specialized heterochromatin at the ends of linear chromosomes. Telomeres are crucial for maintaining genome stability and play important roles in cellular senescence and tumor biology. Six core proteins-TRF1, TRF2, TIN2, POT1, TPP1 and Rap1 (termed the telosome or shelterin complex)-regulate telomere structure and function. One of these proteins, TIN2, regulates telomere length and structure indirectly by interacting with TRF1, TRF2 and TPP1, but no direct function has been attributed to TIN2. Here we present evidence for a TIN2 isoform (TIN2L) that differs from the originally described TIN2 isoform (TIN2S) in two ways: TIN2L contains an additional 97 amino acids, and TIN2L associates strongly with the nuclear matrix. Stringent salt and detergent conditions failed to extract TIN2L from the nuclear matrix, despite removing other telomere components, including TIN2S. In human mammary epithelial cells, each isoform showed a distinct nuclear distribution both as a function of cell cycle position and telomere length. Our results suggest a dual role for TIN2 in mediating the function of the shelterin complex and tethering telomeres to the nuclear matrix.
Figures


Comment in
-
The long and short of it: a new isoform of TIN2 in the nuclear matrix.Cell Cycle. 2009 Mar 15;8(6):797-8. doi: 10.4161/cc.8.6.8337. Epub 2009 Mar 15. Cell Cycle. 2009. PMID: 19279399 No abstract available.
Similar articles
-
The C-Terminal Extension Unique to the Long Isoform of the Shelterin Component TIN2 Enhances Its Interaction with TRF2 in a Phosphorylation- and Dyskeratosis Congenita Cluster-Dependent Fashion.Mol Cell Biol. 2018 May 29;38(12):e00025-18. doi: 10.1128/MCB.00025-18. Print 2018 Jun 15. Mol Cell Biol. 2018. PMID: 29581185 Free PMC article.
-
TIN2 is an architectural protein that facilitates TRF2-mediated trans- and cis-interactions on telomeric DNA.Nucleic Acids Res. 2021 Dec 16;49(22):13000-13018. doi: 10.1093/nar/gkab1142. Nucleic Acids Res. 2021. PMID: 34883513 Free PMC article.
-
TRF2-tethered TIN2 can mediate telomere protection by TPP1/POT1.Mol Cell Biol. 2014 Apr;34(7):1349-62. doi: 10.1128/MCB.01052-13. Epub 2014 Jan 27. Mol Cell Biol. 2014. PMID: 24469404 Free PMC article.
-
Shelterin: the protein complex that shapes and safeguards human telomeres.Genes Dev. 2005 Sep 15;19(18):2100-10. doi: 10.1101/gad.1346005. Genes Dev. 2005. PMID: 16166375 Review.
-
Shelterin proteins and cancer.Asian Pac J Cancer Prev. 2015;16(8):3085-90. doi: 10.7314/apjcp.2015.16.8.3085. Asian Pac J Cancer Prev. 2015. PMID: 25921101 Review.
Cited by
-
The C-Terminal Extension Unique to the Long Isoform of the Shelterin Component TIN2 Enhances Its Interaction with TRF2 in a Phosphorylation- and Dyskeratosis Congenita Cluster-Dependent Fashion.Mol Cell Biol. 2018 May 29;38(12):e00025-18. doi: 10.1128/MCB.00025-18. Print 2018 Jun 15. Mol Cell Biol. 2018. PMID: 29581185 Free PMC article.
-
The Telomere/Telomerase System in Chronic Inflammatory Diseases. Cause or Effect?Genes (Basel). 2016 Sep 3;7(9):60. doi: 10.3390/genes7090060. Genes (Basel). 2016. PMID: 27598205 Free PMC article. Review.
-
Nuclear envelope attachment of telomeres limits TERRA and telomeric rearrangements in quiescent fission yeast cells.Nucleic Acids Res. 2020 Apr 6;48(6):3029-3041. doi: 10.1093/nar/gkaa043. Nucleic Acids Res. 2020. PMID: 31980821 Free PMC article.
-
TIN2 is an architectural protein that facilitates TRF2-mediated trans- and cis-interactions on telomeric DNA.Nucleic Acids Res. 2021 Dec 16;49(22):13000-13018. doi: 10.1093/nar/gkab1142. Nucleic Acids Res. 2021. PMID: 34883513 Free PMC article.
-
Exon-centric regulation of ATM expression is population-dependent and amenable to antisense modification by pseudoexon targeting.Sci Rep. 2016 Jan 6;6:18741. doi: 10.1038/srep18741. Sci Rep. 2016. PMID: 26732650 Free PMC article.
References
-
- Reaper PM, di Fagagna F, Jackson SP. Activation of the DNA damage response by telomere attrition: a passage to cellular senescence. Cell Cycle. 2004;3:543–546. - PubMed
-
- Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:27–31. - PubMed
-
- Hornsby PJ. Senescence as an anticancer mechanism. J Clin Oncol. 2007;25:1852–1857. - PubMed
-
- Campisi J. Senescent cells, tumor suppression and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG017242-100003/AG/NIA NIH HHS/United States
- R37 AG009909-17/AG/NIA NIH HHS/United States
- R37 AG009909-16/AG/NIA NIH HHS/United States
- AG09909/AG/NIA NIH HHS/United States
- AG017242/AG/NIA NIH HHS/United States
- R37 AG009909/AG/NIA NIH HHS/United States
- P01 AG017242-080003/AG/NIA NIH HHS/United States
- P01 AG017242-090003/AG/NIA NIH HHS/United States
- R37 AG009909-18/AG/NIA NIH HHS/United States
- P01 AG017242/AG/NIA NIH HHS/United States
- R37 AG009909-19/AG/NIA NIH HHS/United States
- R56 AG009909/AG/NIA NIH HHS/United States
- P01 AG017242-070003/AG/NIA NIH HHS/United States
- P01 AG017242-060003/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
