An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles
- PMID: 19217333
- PMCID: PMC2696186
- DOI: 10.1016/j.molcel.2009.01.026
An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles
Abstract
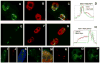
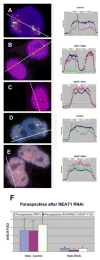
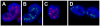
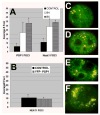
NEAT1 RNA, a highly abundant 4 kb ncRNA, is retained in nuclei in approximately 10 to 20 large foci that we show are completely coincident with paraspeckles, nuclear domains implicated in mRNA nuclear retention. Depletion of NEAT1 RNA via RNAi eradicates paraspeckles, suggesting that it controls sequestration of the paraspeckle proteins PSP1 and p54, factors linked to A-I editing. Unlike overexpression of PSP1, NEAT1 overexpression increases paraspeckle number, and paraspeckles emanate exclusively from the NEAT1 transcription site. The PSP-1 RNA binding domain is required for its colocalization with NEAT1 RNA in paraspeckles, and biochemical analyses support that NEAT1 RNA binds with paraspeckle proteins. Unlike other nuclear-retained RNAs, NEAT1 RNA is not A-I edited, consistent with a structural role in paraspeckles. Collectively, results demonstrate that NEAT1 functions as an essential structural determinant of paraspeckles, providing a precedent for a ncRNA as the foundation of a nuclear domain.
Figures


Similar articles
-
Direct RNA-RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention.RNA Biol. 2021 Nov;18(11):2016-2027. doi: 10.1080/15476286.2021.1889253. Epub 2021 Mar 15. RNA Biol. 2021. PMID: 33573434 Free PMC article.
-
Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1545. doi: 10.1002/wrna.1545. Epub 2019 May 1. Wiley Interdiscip Rev RNA. 2019. PMID: 31044562 Review.
-
Identification and characterization of a special type of subnuclear structure: AGGF1-coated paraspeckles.FASEB J. 2022 Jun;36(6):e22366. doi: 10.1096/fj.202101690RR. FASEB J. 2022. PMID: 35608889
-
Paraspeckles: nuclear bodies built on long noncoding RNA.J Cell Biol. 2009 Sep 7;186(5):637-44. doi: 10.1083/jcb.200906113. Epub 2009 Aug 31. J Cell Biol. 2009. PMID: 19720872 Free PMC article. Review.
-
ARS2 Regulates Nuclear Paraspeckle Formation through 3'-End Processing and Stability of NEAT1 Long Noncoding RNA.Mol Cell Biol. 2020 Jan 30;40(4):e00269-19. doi: 10.1128/MCB.00269-19. Print 2020 Jan 30. Mol Cell Biol. 2020. PMID: 31818879 Free PMC article.
Cited by
-
Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer.Nat Struct Mol Biol. 2013 Jul;20(7):908-13. doi: 10.1038/nsmb.2591. Epub 2013 Jun 2. Nat Struct Mol Biol. 2013. PMID: 23728290 Free PMC article.
-
The regulation of mammalian mRNA transcription by lncRNAs: recent discoveries and current concepts.Epigenomics. 2013 Feb;5(1):95-102. doi: 10.2217/epi.12.69. Epigenomics. 2013. PMID: 23414324 Free PMC article.
-
LncRNAs divide and rule: The master regulators of phase separation.Front Genet. 2022 Aug 10;13:930792. doi: 10.3389/fgene.2022.930792. eCollection 2022. Front Genet. 2022. PMID: 36035193 Free PMC article. Review.
-
Long Intergenic Non-Coding RNAs: Novel Drivers of Human Lymphocyte Differentiation.Front Immunol. 2015 Apr 15;6:175. doi: 10.3389/fimmu.2015.00175. eCollection 2015. Front Immunol. 2015. PMID: 25926836 Free PMC article. Review.
-
NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma.Cell Mol Life Sci. 2021 Mar;78(5):2213-2230. doi: 10.1007/s00018-020-03632-6. Epub 2020 Sep 10. Cell Mol Life Sci. 2021. PMID: 32914209 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- HD007439/HD/NICHD NIH HHS/United States
- R01 DC003263/DC/NIDCD NIH HHS/United States
- GM53234/GM/NIGMS NIH HHS/United States
- GM068711/GM/NIGMS NIH HHS/United States
- T32 HD007439/HD/NICHD NIH HHS/United States
- R01 GM068138-04/GM/NIGMS NIH HHS/United States
- R01 GM053234/GM/NIGMS NIH HHS/United States
- R01 GM068138/GM/NIGMS NIH HHS/United States
- GM68138/GM/NIGMS NIH HHS/United States
- R01 GM053234-07/GM/NIGMS NIH HHS/United States
- DC003263/DC/NIDCD NIH HHS/United States
- R01 GM068711/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

