Landscape Phage as a Molecular Recognition Interface for Detection Devices
- PMID: 19190724
- PMCID: PMC2565273
- DOI: 10.1016/j.mejo.2006.11.007
Landscape Phage as a Molecular Recognition Interface for Detection Devices
Abstract
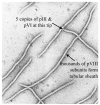
Filamentous phages are thread-shaped bacterial viruses. Their outer coat is a tube formed by thousands equal copies of the major coat protein pVIII. Libraries of random peptides fused to pVIII domains were used for selection of phages probes specific for a panel of test antigens and biological threat agents. Because the viral carrier in the phage borne bio-selective probes is infective, they can be cloned individually and propagated indefinitely without needs of their chemical synthesis or reconstructing. As a new bioselective material, landscape phages combine unique characteristics of affinity reagents and self assembling proteins. Biorecognition layers formed by the phage-derived probes bind biological agents with high affinity and specificity and generate detectable signals in analytical platforms. The performance of phage-derived materials as biorecognition interface was illustrated by detection of Bacillus anthracis spores and Salmonella typhimurium cells. With further refinement, the phage-derived analytical platforms for detecting and monitoring of numerous threat agents may be developed, since phage interface against any bacteria, virus or toxin may be readily selected from the landscape phage libraries. As an interface in the field-use detectors, they may be superior to antibodies, since they are inexpensive, highly specific and strong binders, resistant to high temperatures and environmental stresses.
Figures



Similar articles
-
Phage display for detection of biological threat agents.J Microbiol Methods. 2003 May;53(2):253-62. doi: 10.1016/s0167-7012(03)00029-0. J Microbiol Methods. 2003. PMID: 12654496 Review.
-
Diagnostic probes for Bacillus anthracis spores selected from a landscape phage library.Clin Chem. 2004 Oct;50(10):1899-906. doi: 10.1373/clinchem.2004.038018. Epub 2004 Aug 12. Clin Chem. 2004. PMID: 15308600
-
Specific and selective probes for Pseudomonas aeruginosa from phage-displayed random peptide libraries.Biosens Bioelectron. 2008 Feb 28;23(7):1137-44. doi: 10.1016/j.bios.2007.11.001. Epub 2007 Nov 9. Biosens Bioelectron. 2008. PMID: 18068970
-
Landscape Phage: Evolution from Phage Display to Nanobiotechnology.Viruses. 2018 Jun 7;10(6):311. doi: 10.3390/v10060311. Viruses. 2018. PMID: 29880747 Free PMC article. Review.
-
Homodimeric peptides displayed by the major coat protein of filamentous phage.J Mol Biol. 2000 Jul 7;300(2):307-20. doi: 10.1006/jmbi.2000.3850. J Mol Biol. 2000. PMID: 10873467
Cited by
-
Real-time analysis of dual-display phage immobilization and autoantibody screening using quartz crystal microbalance with dissipation monitoring.Int J Nanomedicine. 2015 Aug 19;10:5237-47. doi: 10.2147/IJN.S84800. eCollection 2015. Int J Nanomedicine. 2015. PMID: 26316752 Free PMC article.
-
Combinatorial synthesis and screening of cancer cell-specific nanomedicines targeted via phage fusion proteins.Front Microbiol. 2015 Jun 23;6:628. doi: 10.3389/fmicb.2015.00628. eCollection 2015. Front Microbiol. 2015. PMID: 26157433 Free PMC article.
-
Nanoscale bacteriophage biosensors beyond phage display.Int J Nanomedicine. 2013;8:3917-25. doi: 10.2147/IJN.S51894. Epub 2013 Oct 10. Int J Nanomedicine. 2013. PMID: 24143096 Free PMC article. Review.
-
Viruses Masquerading as Antibodies in Biosensors: The Development of the Virus BioResistor.Acc Chem Res. 2020 Oct 20;53(10):2384-2394. doi: 10.1021/acs.accounts.0c00474. Epub 2020 Oct 1. Acc Chem Res. 2020. PMID: 33001632 Free PMC article.
-
First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora.Microorganisms. 2020 Nov 10;8(11):1762. doi: 10.3390/microorganisms8111762. Microorganisms. 2020. PMID: 33182526 Free PMC article.
References
-
- Cunningham AJ. Introduction to Bioanalytical Sensors. New york: John Wiley & Sons, Inc.; 1998. p. 418.
-
- Gizeli E, Lowe CR, editors. Biomolecular sensors. Taylor & Francis; London, New York: 2002. p. 322.
-
- Goodchild S, Love T, Hopkins N, Mayers C. Engineering antibodies for biosensor technologies. Adv Appl Microbiol. 2006;58:185–226. - PubMed
-
- Cooper J, Cass T, editors. Biosensors. Second. University Press; Oxford, New York: 2004. p. 251.
-
- Rider TH, Petrovick MS, Nargi FE, Harper JD, Schwoebel ED, Mathews RH, Blanchard DJ, Bortolin LT, Young AM, Chen J, Hollis MA. A B cell-based sensor for rapid identification of pathogens. Science. 2003;301(5630):213–215. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
