Circular dichroism and conformational polymorphism of DNA
- PMID: 19190094
- PMCID: PMC2665218
- DOI: 10.1093/nar/gkp026
Circular dichroism and conformational polymorphism of DNA
Abstract
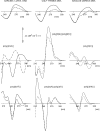
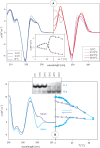
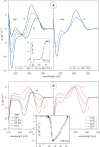
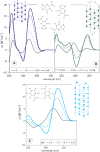
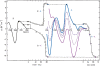
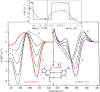
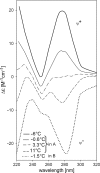
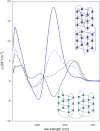
Here we review studies that provided important information about conformational properties of DNA using circular dichroic (CD) spectroscopy. The conformational properties include the B-family of structures, A-form, Z-form, guanine quadruplexes, cytosine quadruplexes, triplexes and other less characterized structures. CD spectroscopy is extremely sensitive and relatively inexpensive. This fast and simple method can be used at low- as well as high-DNA concentrations and with short- as well as long-DNA molecules. The samples can easily be titrated with various agents to cause conformational isomerizations of DNA. The course of detected CD spectral changes makes possible to distinguish between gradual changes within a single DNA conformation and cooperative isomerizations between discrete structural states. It enables measuring kinetics of the appearance of particular conformers and determination of their thermodynamic parameters. In careful hands, CD spectroscopy is a valuable tool for mapping conformational properties of particular DNA molecules. Due to its numerous advantages, CD spectroscopy significantly participated in all basic conformational findings on DNA.
Figures

Similar articles
-
Conformational changes in DNA upon ligand binding monitored by circular dichroism.Int J Mol Sci. 2012;13(3):3394-3413. doi: 10.3390/ijms13033394. Epub 2012 Mar 12. Int J Mol Sci. 2012. PMID: 22489158 Free PMC article. Review.
-
Circular dichroism and guanine quadruplexes.Methods. 2012 May;57(1):64-75. doi: 10.1016/j.ymeth.2012.03.011. Epub 2012 Mar 17. Methods. 2012. PMID: 22450044 Review.
-
CD Study of the G-Quadruplex Conformation.Methods Mol Biol. 2019;2035:25-44. doi: 10.1007/978-1-4939-9666-7_2. Methods Mol Biol. 2019. PMID: 31444742
-
Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes.Chirality. 2012 Sep;24(9):691-8. doi: 10.1002/chir.22064. Epub 2012 Jun 14. Chirality. 2012. PMID: 22696273 Review.
-
Circular Dichroism of DNA G-Quadruplexes: Combining Modeling and Spectroscopy To Unravel Complex Structures.J Phys Chem B. 2016 Mar 31;120(12):3113-21. doi: 10.1021/acs.jpcb.6b00634. Epub 2016 Mar 17. J Phys Chem B. 2016. PMID: 26943487
Cited by
-
Conformational changes in DNA upon ligand binding monitored by circular dichroism.Int J Mol Sci. 2012;13(3):3394-3413. doi: 10.3390/ijms13033394. Epub 2012 Mar 12. Int J Mol Sci. 2012. PMID: 22489158 Free PMC article. Review.
-
Oligoarginine peptides slow strand annealing and assist non-enzymatic RNA replication.Nat Chem. 2016 Oct;8(10):915-21. doi: 10.1038/nchem.2551. Epub 2016 Jun 27. Nat Chem. 2016. Retraction in: Nat Chem. 2017 Nov 23;9(12):1286. doi: 10.1038/nchem.2885 PMID: 27657866 Free PMC article. Retracted.
-
Anti-Inflammatory and Antioxidant Pyrrolo[3,4-d]pyridazinone Derivatives Interact with DNA and Bind to Plasma Proteins-Spectroscopic and In Silico Studies.Int J Mol Sci. 2024 Feb 1;25(3):1784. doi: 10.3390/ijms25031784. Int J Mol Sci. 2024. PMID: 38339061 Free PMC article.
-
Oligonucleotide models of telomeric DNA and RNA form a Hybrid G-quadruplex structure as a potential component of telomeres.J Biol Chem. 2012 Dec 7;287(50):41787-96. doi: 10.1074/jbc.M112.342030. Epub 2012 Sep 25. J Biol Chem. 2012. PMID: 23012368 Free PMC article.
-
Detection of G-quadruplex DNA using primer extension as a tool.PLoS One. 2015 Mar 23;10(3):e0119722. doi: 10.1371/journal.pone.0119722. eCollection 2015. PLoS One. 2015. PMID: 25799152 Free PMC article.
References
-
- Neidle S. Nucleic Acid Structure. New York: Oxford University Press Inc.; 1999.
-
- Mirkin SM. Discovery of alternative DNA structures: a heroic decade (1979-1989) Front. Biosci. 2008;13:1064–1071. - PubMed
-
- Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. - PubMed
-
- Nakanishi K, Berova N, Woody RW. Circular Dichroism Principles and Applications. New York: VCH Publishers Inc.; 1991.
-
- Woody RW. Circular dichroism. Methods Enzymol. 1995;246:34–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

