The fascinating world of RNA interference
- PMID: 19173032
- PMCID: PMC2631224
- DOI: 10.7150/ijbs.5.97
The fascinating world of RNA interference
Abstract
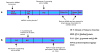
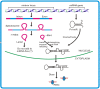
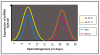
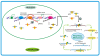
Micro- and short-interfering RNAs represent small RNA family that are recognized as critical regulatory species across the eukaryotes. Recent high-throughput sequencing have revealed two more hidden players of the cellular small RNA pool. Reported in mammals and Caenorhabditis elegans respectively, these new small RNAs are named piwi-interacting RNAs (piRNAs) and 21U-RNAs. Moreover, small RNAs including miRNAs have been identified in unicellular alga Chlamydomonas reinhardtii, redefining the earlier concept of multi-cellularity restricted presence of these molecules. The discovery of these species of small RNAs has allowed us to understand better the usage of genome and the number of genes present but also have complicated the situation in terms of biochemical attributes and functional genesis of these molecules. Nonetheless, these new pools of knowledge have opened up avenues for unraveling the finer details of the small RNA mediated pathways.
Keywords: 21-U RNA; argonaute; dicer; miRNA; mirtron.; piRNA; siRNA.
Conflict of interest statement
Conflict of Interest: The authors have declared that no conflict of interest exists.
Figures


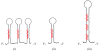

Similar articles
-
A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii.Genes Dev. 2007 May 15;21(10):1190-203. doi: 10.1101/gad.1543507. Epub 2007 Apr 30. Genes Dev. 2007. PMID: 17470535 Free PMC article.
-
PID-1 is a novel factor that operates during 21U-RNA biogenesis in Caenorhabditis elegans.Genes Dev. 2014 Apr 1;28(7):683-8. doi: 10.1101/gad.238220.114. Genes Dev. 2014. PMID: 24696453 Free PMC article.
-
High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes.Genome Res. 2013 Mar;23(3):497-508. doi: 10.1101/gr.149112.112. Epub 2013 Jan 30. Genome Res. 2013. PMID: 23363624 Free PMC article.
-
Diverse small non-coding RNAs in RNA interference pathways.Methods Mol Biol. 2011;764:169-82. doi: 10.1007/978-1-61779-188-8_11. Methods Mol Biol. 2011. PMID: 21748640 Review.
-
Argonaute proteins: mediators of RNA silencing.Mol Cell. 2007 Jun 8;26(5):611-23. doi: 10.1016/j.molcel.2007.05.001. Mol Cell. 2007. PMID: 17560368 Review.
Cited by
-
Effects of HBV Genetic Variability on RNAi Strategies.Hepat Res Treat. 2011;2011:367908. doi: 10.1155/2011/367908. Epub 2011 Jul 2. Hepat Res Treat. 2011. PMID: 21760994 Free PMC article.
-
Viral MicroRNAs: Interfering the Interferon Signaling.Curr Pharm Des. 2020;26(4):446-454. doi: 10.2174/1381612826666200109181238. Curr Pharm Des. 2020. PMID: 31924149 Free PMC article. Review.
-
Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA.J Biol Chem. 2013 Jul 5;288(27):20014-33. doi: 10.1074/jbc.M112.438895. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661700 Free PMC article.
-
miR-223-3p Inhibits Antigen Endocytosis and Presentation and Promotes the Tolerogenic Potential of Dendritic Cells through Targeting Mannose Receptor Signaling and Rhob.J Immunol Res. 2020 Jun 18;2020:1379458. doi: 10.1155/2020/1379458. eCollection 2020. J Immunol Res. 2020. PMID: 32656268 Free PMC article.
-
Long Non-Coding RNAs and Their Potential Roles in the Vector-Host-Pathogen Triad.Life (Basel). 2021 Jan 14;11(1):56. doi: 10.3390/life11010056. Life (Basel). 2021. PMID: 33466803 Free PMC article. Review.
References
-
- Fire A, Xu S, Montgomery M et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. - PubMed
-
- Hamilton A, Baulcombe D. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–2. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

