High-resolution DNA-binding specificity analysis of yeast transcription factors
- PMID: 19158363
- PMCID: PMC2665775
- DOI: 10.1101/gr.090233.108
High-resolution DNA-binding specificity analysis of yeast transcription factors
Abstract
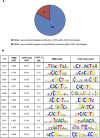
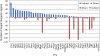
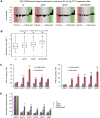
Transcription factors (TFs) regulate the expression of genes through sequence-specific interactions with DNA-binding sites. However, despite recent progress in identifying in vivo TF binding sites by microarray readout of chromatin immunoprecipitation (ChIP-chip), nearly half of all known yeast TFs are of unknown DNA-binding specificities, and many additional predicted TFs remain uncharacterized. To address these gaps in our knowledge of yeast TFs and their cis regulatory sequences, we have determined high-resolution binding profiles for 89 known and predicted yeast TFs, over more than 2.3 million gapped and ungapped 8-bp sequences ("k-mers"). We report 50 new or significantly different direct DNA-binding site motifs for yeast DNA-binding proteins and motifs for eight proteins for which only a consensus sequence was previously known; in total, this corresponds to over a 50% increase in the number of yeast DNA-binding proteins with experimentally determined DNA-binding specificities. Among other novel regulators, we discovered proteins that bind the PAC (Polymerase A and C) motif (GATGAG) and regulate ribosomal RNA (rRNA) transcription and processing, core cellular processes that are constituent to ribosome biogenesis. In contrast to earlier data types, these comprehensive k-mer binding data permit us to consider the regulatory potential of genomic sequence at the individual word level. These k-mer data allowed us to reannotate in vivo TF binding targets as direct or indirect and to examine TFs' potential effects on gene expression in approximately 1,700 environmental and cellular conditions. These approaches could be adapted to identify TFs and cis regulatory elements in higher eukaryotes.
Figures


Similar articles
-
Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights.Genome Biol. 2011 Dec 21;12(12):R125. doi: 10.1186/gb-2011-12-12-r125. Genome Biol. 2011. PMID: 22189060 Free PMC article.
-
Identifying combinatorial regulation of transcription factors and binding motifs.Genome Biol. 2004;5(8):R56. doi: 10.1186/gb-2004-5-8-r56. Epub 2004 Jul 28. Genome Biol. 2004. PMID: 15287978 Free PMC article.
-
Distinguishing direct versus indirect transcription factor-DNA interactions.Genome Res. 2009 Nov;19(11):2090-100. doi: 10.1101/gr.094144.109. Epub 2009 Aug 3. Genome Res. 2009. PMID: 19652015 Free PMC article.
-
Transcription factor-DNA binding: beyond binding site motifs.Curr Opin Genet Dev. 2017 Apr;43:110-119. doi: 10.1016/j.gde.2017.02.007. Epub 2017 Mar 27. Curr Opin Genet Dev. 2017. PMID: 28359978 Free PMC article. Review.
-
Transcriptional networks: reverse-engineering gene regulation on a global scale.Curr Opin Microbiol. 2004 Dec;7(6):638-46. doi: 10.1016/j.mib.2004.10.009. Curr Opin Microbiol. 2004. PMID: 15556037 Review.
Cited by
-
Dynamic changes in nucleosome occupancy are not predictive of gene expression dynamics but are linked to transcription and chromatin regulators.Mol Cell Biol. 2012 May;32(9):1645-53. doi: 10.1128/MCB.06170-11. Epub 2012 Feb 21. Mol Cell Biol. 2012. PMID: 22354995 Free PMC article.
-
Mapping functional transcription factor networks from gene expression data.Genome Res. 2013 Aug;23(8):1319-28. doi: 10.1101/gr.150904.112. Epub 2013 May 1. Genome Res. 2013. PMID: 23636944 Free PMC article.
-
Unraveling determinants of transcription factor binding outside the core binding site.Genome Res. 2015 Jul;25(7):1018-29. doi: 10.1101/gr.185033.114. Epub 2015 Mar 11. Genome Res. 2015. PMID: 25762553 Free PMC article.
-
A compendium of DNA-binding specificities of transcription factors in Pseudomonas syringae.Nat Commun. 2020 Oct 2;11(1):4947. doi: 10.1038/s41467-020-18744-7. Nat Commun. 2020. PMID: 33009392 Free PMC article.
-
The yin and yang of yeast transcription: elements of a global feedback system between metabolism and chromatin.PLoS One. 2012;7(6):e37906. doi: 10.1371/journal.pone.0037906. Epub 2012 Jun 7. PLoS One. 2012. PMID: 22685547 Free PMC article.
References
-
- Angus-Hill M.L., Schlichter A., Roberts D., Erdjument-Bromage H., Tempst P., Cairns B.R. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell. 2001;7:741–751. - PubMed
-
- Beer M.A., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
