Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells
- PMID: 19151416
- PMCID: PMC2689770
- DOI: 10.1074/mcp.M800287-MCP200
Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells
Abstract
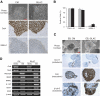
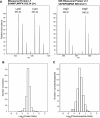
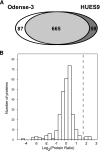
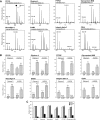
Stable isotope labeling by amino acids in cell culture (SILAC) is a powerful quantitative proteomics platform for comprehensive characterization of complex biological systems. However, the potential of SILAC-based approaches has not been fully utilized in human embryonic stem cell (hESC) research mainly because of the complex nature of hESC culture conditions. Here we describe complete SILAC labeling of hESCs with fully preserved pluripotency, self-renewal capabilities, and overall proteome status that was quantitatively analyzed to a depth of 1556 proteins and 527 phosphorylation events. SILAC-labeled hESCs appear to be perfectly suitable for functional studies, and we exploited a SILAC-based proteomics strategy for discovery of hESC-specific surface markers. We determined and quantitatively compared the membrane proteomes of the self-renewing versus differentiating cells of two distinct human embryonic stem cell lines. Of the 811 identified membrane proteins, six displayed significantly higher expression levels in the undifferentiated state compared with differentiating cells. This group includes the established marker CD133/Prominin-1 as well as novel candidates for hESC surface markers: Glypican-4, Neuroligin-4, ErbB2, receptor-type tyrosine-protein phosphatase zeta (PTPRZ), and Glycoprotein M6B. Our study also revealed 17 potential markers of hESC differentiation as their corresponding protein expression levels displayed a dramatic increase in differentiated embryonic stem cell populations.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Prevention of amino acid conversion in SILAC experiments with embryonic stem cells.Mol Cell Proteomics. 2008 Sep;7(9):1587-97. doi: 10.1074/mcp.M800113-MCP200. Epub 2008 May 16. Mol Cell Proteomics. 2008. PMID: 18487603 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7 PMID: 38260445 Free PMC article. Updated. Preprint.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Plasma membrane proteomics of tumor spheres identify CD166 as a novel marker for cancer stem-like cells in head and neck squamous cell carcinoma.Mol Cell Proteomics. 2013 Nov;12(11):3271-84. doi: 10.1074/mcp.M112.025460. Epub 2013 Jul 31. Mol Cell Proteomics. 2013. PMID: 23903875 Free PMC article.
-
Cleavage of E-cadherin and β-catenin by calpain affects Wnt signaling and spheroid formation in suspension cultures of human pluripotent stem cells.Mol Cell Proteomics. 2014 Apr;13(4):990-1007. doi: 10.1074/mcp.M113.033423. Epub 2014 Jan 30. Mol Cell Proteomics. 2014. PMID: 24482122 Free PMC article.
-
Stable isotope dilution mass spectrometry for membrane transporter quantitation.AAPS J. 2013 Oct;15(4):1222-31. doi: 10.1208/s12248-013-9529-8. AAPS J. 2013. PMID: 24022320 Free PMC article. Review.
-
The quest of cell surface markers for stem cell therapy.Cell Mol Life Sci. 2021 Jan;78(2):469-495. doi: 10.1007/s00018-020-03602-y. Epub 2020 Jul 24. Cell Mol Life Sci. 2021. PMID: 32710154 Free PMC article. Review.
-
Quantitative top-down proteomics of SILAC labeled human embryonic stem cells.J Am Soc Mass Spectrom. 2010 Jun;21(6):879-89. doi: 10.1016/j.jasms.2010.01.031. Epub 2010 Feb 6. J Am Soc Mass Spectrom. 2010. PMID: 20199872
References
-
- Lerou, P. H., and Daley, G. Q. ( 2005) Therapeutic potential of embryonic stem cells. Blood Rev. 19, 321–331 - PubMed
-
- Ohtsuka, S., and Dalton, S. ( 2008) Molecular and biological properties of pluripotent embryonic stem cells. Gene Ther. 15, 74–81 - PubMed
-
- Hanna, J., Markoulaki, S., Schorderet, P., Carey, B. W., Beard, C., Wernig, M., Creyghton, M. P., Steine, E. J., Cassady, J. P., Foreman, R., Lengner, C. J., Dausman, J. A., and Jaenisch, R. ( 2008) Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133, 250–264 - PMC - PubMed
-
- Nakagawa, M., Koyanagi, M., Tanabe, K., Takahashi, K., Ichisaka, T., Aoi, T., Okita, K., Mochiduki, Y., Takizawa, N., and Yamanaka, S. ( 2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

