E4orf1 limits the oncolytic potential of the E1B-55K deletion mutant adenovirus
- PMID: 19129452
- PMCID: PMC2648266
- DOI: 10.1128/JVI.01972-08
E4orf1 limits the oncolytic potential of the E1B-55K deletion mutant adenovirus
Abstract
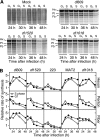
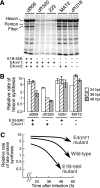
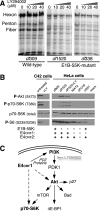
Clinical trials have shown oncolytic adenoviruses to be tumor selective with minimal toxicity toward normal tissue. The virus ONYX-015, in which the gene encoding the early region 1B 55-kDa (E1B-55K) protein is deleted, has been most effective when used in combination with either chemotherapy or radiation therapy. Therefore, improving the oncolytic nature of tumor-selective adenoviruses remains an important objective for improving this form of cancer therapy. Cells infected during the G(1) phase of the cell cycle with the E1B-55K deletion mutant virus exhibit a reduced rate of viral late protein synthesis, produce fewer viral progeny, and are less efficiently killed than cells infected during the S phase. Here we demonstrate that the G(1) restriction imposed on the E1B-55K deletion mutant virus is due to the viral oncogene encoded by open reading frame 1 of early region 4 (E4orf1). E4orf1 has been reported to signal through the phosphatidylinositol 3'-kinase pathway leading to the activation of Akt, mTOR, and p70 S6K. Evidence presented here shows that E4orf1 may also induce the phosphorylation of Akt and p70 S6K in a manner that depends on Rac1 and its guanine nucleotide exchange factor Tiam1. Accordingly, agents that have been reported to disrupt the Tiam1-Rac1 interaction or to prevent phosphorylation of the ribosomal S6 kinase partially alleviated the E4orf1 restriction to late viral protein synthesis and enhanced tumor cell killing by the E1B-55K mutant virus. These results demonstrate that E4orf1 limits the oncolytic nature of a conditionally replicating adenovirus such as ONYX-015. The therapeutic value of similar oncolytic adenoviruses may be improved by abrogating E4orf1 function.
Figures

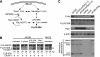

Similar articles
-
The human adenovirus PI3K-Akt activator E4orf1 is targeted by the tumor suppressor p53.J Virol. 2024 Apr 16;98(4):e0170123. doi: 10.1128/jvi.01701-23. Epub 2024 Mar 7. J Virol. 2024. PMID: 38451084 Free PMC article.
-
The adenovirus E1B 55-kilodalton and E4 open reading frame 6 proteins limit phosphorylation of eIF2alpha during the late phase of infection.J Virol. 2009 Oct;83(19):9970-82. doi: 10.1128/JVI.01113-09. Epub 2009 Jul 15. J Virol. 2009. PMID: 19605483 Free PMC article.
-
Functional interactions of antiapoptotic proteins and tumor necrosis factor in the context of a replication-competent adenovirus.Gene Ther. 2005 Sep;12(17):1333-46. doi: 10.1038/sj.gt.3302555. Gene Ther. 2005. PMID: 15920462
-
Oncolytic Replication of E1b-Deleted Adenoviruses.Viruses. 2015 Nov 6;7(11):5767-79. doi: 10.3390/v7112905. Viruses. 2015. PMID: 26561828 Free PMC article. Review.
-
The biology of the adenovirus E1B 55K protein.FEBS Lett. 2019 Dec;593(24):3504-3517. doi: 10.1002/1873-3468.13694. Epub 2019 Dec 8. FEBS Lett. 2019. PMID: 31769868 Review.
Cited by
-
Impact of Autophagy in Oncolytic Adenoviral Therapy for Cancer.Int J Mol Sci. 2017 Jul 10;18(7):1479. doi: 10.3390/ijms18071479. Int J Mol Sci. 2017. PMID: 28698504 Free PMC article. Review.
-
Tinkering with translation: protein synthesis in virus-infected cells.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012351. doi: 10.1101/cshperspect.a012351. Cold Spring Harb Perspect Biol. 2013. PMID: 23209131 Free PMC article. Review.
-
The human adenovirus PI3K-Akt activator E4orf1 is targeted by the tumor suppressor p53.J Virol. 2024 Apr 16;98(4):e0170123. doi: 10.1128/jvi.01701-23. Epub 2024 Mar 7. J Virol. 2024. PMID: 38451084 Free PMC article.
-
Adenovirus replaces mitotic checkpoint controls.J Virol. 2015 May;89(9):5083-96. doi: 10.1128/JVI.00213-15. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694601 Free PMC article.
-
Effects of the deletion of early region 4 (E4) open reading frame 1 (orf1), orf1-2, orf1-3 and orf1-4 on virus-host cell interaction, transgene expression, and immunogenicity of replicating adenovirus HIV vaccine vectors.PLoS One. 2013 Oct 15;8(10):e76344. doi: 10.1371/journal.pone.0076344. eCollection 2013. PLoS One. 2013. PMID: 24143187 Free PMC article.
References
-
- Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156107-121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

