MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis
- PMID: 19106625
- PMCID: PMC3972804
- DOI: 10.4161/rna.6.1.7534
MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis
Abstract
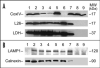
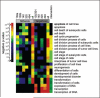
MicroRNAs (miRNAs) are a class of small approximately 22 nt noncoding (nc) RNAs that regulate gene expression post-transcriptionally by direct binding to target sites on mRNAs. They comprise more than 1,000 novel species in mammalian cells and exert their function by modulating gene expression through several different mechanisms, including translational inhibition, and/or degradation of target mRNAs. Mitochondria maintain and express their own genome, which is distinct from the nuclear transcriptional and translational apparatus. Thus, they provide a potential site for miRNA mediated post-transcriptional regulation. To determine whether they maintain a unique miRNA population, we examined the miRNA profile from highly purified and RNase treated mitochondria from adult rat liver. Fifteen miRNAs were identified by microarray analysis of which, five were confirmed by TaqMan 5'nuclease assays using rat specific probes. Functional analysis of the miRNAs indicated that they were not targeted to the mitochondrial genome nor were they complementary to nuclear RNAs encoding mitochondrial proteins. Rather, the mitochondria-associated miRNAs appear to be involved in the expression of genes associated with apoptosis, cell proliferation, and differentiation. Given the central role that mitochondria play in apoptosis, the results suggest that they might serve as reservoirs of select miRNAs that may modulate these processes in a coordinate fashion.
Figures

Similar articles
-
The Emerging Role of MitomiRs in the Pathophysiology of Human Disease.Adv Exp Med Biol. 2015;888:123-54. doi: 10.1007/978-3-319-22671-2_8. Adv Exp Med Biol. 2015. PMID: 26663182
-
Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells.J Biol Chem. 2009 Mar 20;284(12):7903-13. doi: 10.1074/jbc.M806920200. Epub 2009 Jan 21. J Biol Chem. 2009. PMID: 19158092 Free PMC article.
-
MicroRNA: past and present.Front Biosci. 2007 Jan 1;12:2316-29. doi: 10.2741/2234. Front Biosci. 2007. PMID: 17127242 Review.
-
Nuclear outsourcing of RNA interference components to human mitochondria.PLoS One. 2011;6(6):e20746. doi: 10.1371/journal.pone.0020746. Epub 2011 Jun 13. PLoS One. 2011. PMID: 21695135 Free PMC article.
-
Function and localization of microRNAs in mammalian cells.Cold Spring Harb Symp Quant Biol. 2006;71:29-38. doi: 10.1101/sqb.2006.71.049. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381277 Review.
Cited by
-
A landscape of mouse mitochondrial small non-coding RNAs.PLoS One. 2024 Jan 2;19(1):e0293644. doi: 10.1371/journal.pone.0293644. eCollection 2024. PLoS One. 2024. PMID: 38165955 Free PMC article.
-
A critical evaluation of neuroprotective and neurodegenerative MicroRNAs in Alzheimer's disease.Biochem Biophys Res Commun. 2017 Feb 19;483(4):1156-1165. doi: 10.1016/j.bbrc.2016.08.067. Epub 2016 Aug 12. Biochem Biophys Res Commun. 2017. PMID: 27524239 Free PMC article. Review.
-
Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis.Biomolecules. 2022 Dec 13;12(12):1863. doi: 10.3390/biom12121863. Biomolecules. 2022. PMID: 36551291 Free PMC article. Review.
-
Mitochondrial epigenetics in aging and cardiovascular diseases.Front Cardiovasc Med. 2023 Jul 13;10:1204483. doi: 10.3389/fcvm.2023.1204483. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37522089 Free PMC article. Review.
-
A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa).BMC Genomics. 2011 Jun 3;12:289. doi: 10.1186/1471-2164-12-289. BMC Genomics. 2011. PMID: 21639890 Free PMC article.
References
-
- Tuschl T. Functional genomics: RNA sets the standard. Nature. 2003;421:220–1. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. - PubMed
-
- Meyers BC, Souret FF, Lu C, Green PJ. Sweating the small stuff: microRNA discovery in plants. Curr Opin Biotechnol. 2006;17:139–46. - PubMed
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
