Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma
- PMID: 19098997
- PMCID: PMC2605622
- DOI: 10.2119/molmed.2008.00110
Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma
Abstract
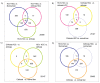
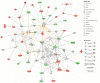
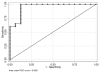
The role of chronic hepatitis C virus (HCV) in the pathogenesis of HCV-associated hepatocellular carcinoma (HCC) remains controversial. To understand the transition from benign to malignant, we studied the gene expression patterns in liver tissues at different stages, including normal, cirrhosis, and different HCC stages. We studied 108 liver tissue samples obtained from 88 distinct patients (41 HCV-cirrhotic tissues, 17 HCV-cirrhotic tissues from patients with HCC, and 47 HCV-HCC tissues). Differentially expressed genes (DEG) were studied by use of high-density oligonucleotide arrays. Among probe sets identified as differentially expressed via the F test, all pairwise comparisons were performed. Cirrhotic tissues with and without concomitant HCC were further evaluated, and a classifier was used to predict whether the tissue type was associated with HCC. Differential expression profiles were analyzed using Interaction Networks and Functional Analysis. Characteristic gene signatures were identified when normal tissue was compared with cirrhosis, cirrhosis with early HCC, and normal with HCC. Pathway analysis classified the cellular and biological functions of the DEG as related to cellular growth and proliferation, cell death and inflammatory disease in cirrhosis; cell death, cell cycle, DNA replication, and immune response in early HCCs; and cell death, cell growth and proliferation, cell cycle, and DNA repair in advanced HCCs. Characteristic gene signatures were identified at different stages of HCV-HCC progression. A set of genes were identified to predict whether the cirrhotic tissue was associated with HCC.
Figures

Similar articles
-
Molecular signatures associated with HCV-induced hepatocellular carcinoma and liver metastasis.PLoS One. 2013;8(2):e56153. doi: 10.1371/journal.pone.0056153. Epub 2013 Feb 18. PLoS One. 2013. PMID: 23441164 Free PMC article.
-
Specific gene-expression profiles of noncancerous liver tissue predict the risk for multicentric occurrence of hepatocellular carcinoma in hepatitis C virus-positive patients.Ann Surg Oncol. 2006 Jul;13(7):947-54. doi: 10.1245/ASO.2006.07.018. Epub 2006 May 23. Ann Surg Oncol. 2006. PMID: 16788756
-
Integrative network analysis identifies key genes and pathways in the progression of hepatitis C virus induced hepatocellular carcinoma.BMC Med Genomics. 2011 Aug 8;4:62. doi: 10.1186/1755-8794-4-62. BMC Med Genomics. 2011. PMID: 21824427 Free PMC article.
-
Molecular pathogenesis of hepatitis C virus-associated hepatocellular carcinoma.Front Biosci. 2007 Jan 1;12:222-33. doi: 10.2741/2060. Front Biosci. 2007. PMID: 17127295 Review.
-
Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer.Oncogene. 2011 Apr 28;30(17):1969-83. doi: 10.1038/onc.2010.594. Epub 2011 Jan 24. Oncogene. 2011. PMID: 21258404 Free PMC article. Review.
Cited by
-
Tumor-suppressing function of caspase-2 requires catalytic site Cys-320 and site Ser-139 in mice.J Biol Chem. 2012 Apr 27;287(18):14792-802. doi: 10.1074/jbc.M112.347625. Epub 2012 Mar 6. J Biol Chem. 2012. PMID: 22396545 Free PMC article.
-
Reconstructing dynamic gene regulatory networks from sample-based transcriptional data.Nucleic Acids Res. 2012 Nov;40(21):10657-67. doi: 10.1093/nar/gks860. Epub 2012 Sep 21. Nucleic Acids Res. 2012. PMID: 23002138 Free PMC article.
-
CD97 serves as a novel biomarker of immune cell infiltration in hepatocellular carcinoma.World J Surg Oncol. 2022 Dec 4;20(1):382. doi: 10.1186/s12957-022-02829-2. World J Surg Oncol. 2022. PMID: 36464675 Free PMC article.
-
Hec1/Ndc80 is overexpressed in human gastric cancer and regulates cell growth.J Gastroenterol. 2014 Mar;49(3):408-18. doi: 10.1007/s00535-013-0809-y. Epub 2013 Apr 17. J Gastroenterol. 2014. PMID: 23591767
-
Krüppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression.PLoS One. 2012;7(8):e43593. doi: 10.1371/journal.pone.0043593. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937066 Free PMC article.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. - PubMed
-
- Tagger A, Donato F, Ribero ML, et al. Case-control study on hepatitis C virus (HCV) as a risk factor for hepatocellular carcinoma: the role of HCV genotypes and the synergism with hepatitis B virus and alcohol. Brescia HCC Study. Int J Cancer. 1999;81:695–699. - PubMed
-
- Tsukuma H, Hiyama T, Tanaka S, Nakao M, Yabuuchi T, Kitamura T, Nakanishi K, et al. Risk factors for hepatocellular carcinoma patients with chronic liver disease. N Engl J Med. 1993;328:1797–1801. - PubMed
-
- El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. - PubMed
-
- Marsh JW, Dvorchik I. Liver organ allocation for hepatocellular carcinoma: are we sure? Liver Transpl. 2003;9:693–696. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 DK069859/DK/NIDDK NIH HHS/United States
- DK62505/DK/NIDDK NIH HHS/United States
- U01 DK062531/DK/NIDDK NIH HHS/United States
- DK62444/DK/NIDDK NIH HHS/United States
- U01 DK062498/DK/NIDDK NIH HHS/United States
- U01 DK062444/DK/NIDDK NIH HHS/United States
- U01 DK062505/DK/NIDDK NIH HHS/United States
- DK62531/DK/NIDDK NIH HHS/United States
- U01 DK062467/DK/NIDDK NIH HHS/United States
- R01DK069859/DK/NIDDK NIH HHS/United States
- DK62498/DK/NIDDK NIH HHS/United States
- U01 DK062496/DK/NIDDK NIH HHS/United States
- DK62494/DK/NIDDK NIH HHS/United States
- DK62467/DK/NIDDK NIH HHS/United States
- DK62496/DK/NIDDK NIH HHS/United States
- U01 DK062494/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

