Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations
- PMID: 19055775
- PMCID: PMC2607301
- DOI: 10.1186/1471-2164-9-581
Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations
Abstract
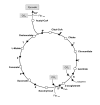
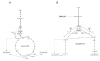
Background: Carbon and nitrogen fixation are essential pathways for autotrophic bacteria living in extreme environments. These bacteria can use carbon dioxide directly from the air as their sole carbon source and can use different sources of nitrogen such as ammonia, nitrate, nitrite, or even nitrogen from the air. To have a better understanding of how these processes occur and to determine how we can make them more efficient, a comparative genomic analysis of three bioleaching bacteria isolated from mine sites in Chile was performed. This study demonstrated that there are important differences in the carbon dioxide and nitrogen fixation mechanisms among bioleaching bacteria that coexist in mining environments.
Results: In this study, we probed that both Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans incorporate CO2 via the Calvin-Benson-Bassham cycle; however, the former bacterium has two copies of the Rubisco type I gene whereas the latter has only one copy. In contrast, we demonstrated that Leptospirillum ferriphilum utilizes the reductive tricarboxylic acid cycle for carbon fixation. Although all the species analyzed in our study can incorporate ammonia by an ammonia transporter, we demonstrated that Acidithiobacillus thiooxidans could also assimilate nitrate and nitrite but only Acidithiobacillus ferrooxidans could fix nitrogen directly from the air.
Conclusion: The current study utilized genomic and molecular evidence to verify carbon and nitrogen fixation mechanisms for three bioleaching bacteria and provided an analysis of the potential regulatory pathways and functional networks that control carbon and nitrogen fixation in these microorganisms.
Figures

Similar articles
-
Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans.BMC Microbiol. 2010 Aug 27;10:229. doi: 10.1186/1471-2180-10-229. BMC Microbiol. 2010. PMID: 20799944 Free PMC article.
-
Expression and activity of the Calvin-Benson-Bassham cycle transcriptional regulator CbbR from Acidithiobacillus ferrooxidans in Ralstonia eutropha.FEMS Microbiol Lett. 2015 Aug;362(15):fnv108. doi: 10.1093/femsle/fnv108. Epub 2015 Jul 6. FEMS Microbiol Lett. 2015. PMID: 26152700
-
Bioleaching of metals from printed wire boards by Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans and their mixture.J Hazard Mater. 2009 Dec 30;172(2-3):1100-5. doi: 10.1016/j.jhazmat.2009.07.102. Epub 2009 Aug 3. J Hazard Mater. 2009. PMID: 19699031
-
Acidithiobacillus thiooxidans and its potential application.Appl Microbiol Biotechnol. 2019 Oct;103(19):7819-7833. doi: 10.1007/s00253-019-10098-5. Epub 2019 Aug 28. Appl Microbiol Biotechnol. 2019. PMID: 31463545 Review.
-
Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments.Environ Microbiol. 2012 Jul;14(7):1597-611. doi: 10.1111/j.1462-2920.2011.02626.x. Epub 2011 Nov 3. Environ Microbiol. 2012. PMID: 22050575 Review.
Cited by
-
Draft genome sequence of the Sulfobacillus thermosulfidooxidans Cutipay strain, an indigenous bacterium isolated from a naturally extreme mining environment in Northern Chile.J Bacteriol. 2012 Nov;194(22):6327-8. doi: 10.1128/JB.01622-12. J Bacteriol. 2012. PMID: 23105067 Free PMC article.
-
Metabolomic study of Chilean biomining bacteria Acidithiobacillus ferrooxidans strain Wenelen and Acidithiobacillus thiooxidans strain Licanantay.Metabolomics. 2013 Feb;9(1):247-257. doi: 10.1007/s11306-012-0443-3. Epub 2012 Jul 21. Metabolomics. 2013. PMID: 23335869 Free PMC article.
-
Carbon fixation by basalt-hosted microbial communities.Front Microbiol. 2015 Sep 7;6:904. doi: 10.3389/fmicb.2015.00904. eCollection 2015. Front Microbiol. 2015. PMID: 26441854 Free PMC article.
-
In Silico Genome-Wide Analysis Reveals the Potential Links Between Core Genome of Acidithiobacillus thiooxidans and Its Autotrophic Lifestyle.Front Microbiol. 2018 Jun 8;9:1255. doi: 10.3389/fmicb.2018.01255. eCollection 2018. Front Microbiol. 2018. PMID: 29937764 Free PMC article.
-
The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer.Front Microbiol. 2013 Feb 21;4:27. doi: 10.3389/fmicb.2013.00027. eCollection 2013. Front Microbiol. 2013. PMID: 23439773 Free PMC article.
References
-
- Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, 2nd, Shah M, Hettich RL, Banfield JF. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. 10.1126/science. 1109070. - PubMed
-
- Valdes J, Pedroso I, Quatrini R, Holmes DS. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into their metabolism and ecophysiology. Hydrometallurgy. 2008;94:180–184. doi: 10.1016/j.hydromet.2008.05.039. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

