Regulation of the phage phi 29 prohead shape and size by the portal vertex
- PMID: 1905079
- PMCID: PMC4167689
- DOI: 10.1016/0042-6822(91)90149-6
Regulation of the phage phi 29 prohead shape and size by the portal vertex
Abstract
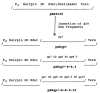
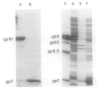
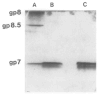
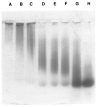
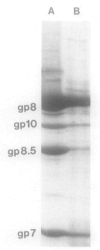
Bacteriophage phi 29 of Bacillus subtilis packages its double-stranded DNA into a preformed prohead during morphogenesis. The prohead is composed of the scaffold protein gp7, the capsid protein pg8, the portal protein gp10, and the dispensable head fiber protein gp8.5. Our objective was to elucidate the phi 29 prohead assembly pathway and to define the factors that determine prohead shape and size. The structural genes of the phi 29 prohead were cloned and expressed in Escherichia coli individually or in combination to study form determination. The scaffold protein was purified from E. coli as a soluble monomer. In vivo and in vitro studies showed that the scaffolding protein interacted with both the portal vertex and capsid proteins. When the scaffold protein interacted only with the capsid protein in vivo, particles were formed with variable size and shape. However, in the presence of the portal vertex protein, particles with uniform size and shape were produced in vivo. SDS-PAGE analysis showed that the latter particles contained the proteins of the scaffold, capsid, head fiber, and portal vertex. These results suggest that the scaffolding protein serves as the linkage between the portal vertex and the capsid proteins, and that the portal vertex plays a crucial role in regulating the size and shape of the prohead.
Figures



Similar articles
-
sRNA of phage phi 29 of Bacillus subtilis mediates DNA packaging of phi 29 proheads assembled in Escherichia coli.Virology. 1991 Nov;185(1):395-400. doi: 10.1016/0042-6822(91)90787-c. Virology. 1991. PMID: 1926784
-
Sequential interactions of structural proteins in phage phi 29 procapsid assembly.J Virol. 1995 Aug;69(8):5024-32. doi: 10.1128/JVI.69.8.5024-5032.1995. J Virol. 1995. PMID: 7609072 Free PMC article.
-
Novel mutants in the 5' upstream region of the portal protein gene 20 overcome a gp40-dependent prohead assembly block in bacteriophage T4.J Mol Biol. 1996 Nov 8;263(4):539-50. doi: 10.1006/jmbi.1996.0597. J Mol Biol. 1996. PMID: 8918937
-
Role of RNA in bacteriophage phi 29 DNA packaging.J Struct Biol. 1990 Jul-Sep;104(1-3):70-4. doi: 10.1016/1047-8477(90)90059-l. J Struct Biol. 1990. PMID: 2150913
-
Form determination of the heads of bacteriophages.Eur J Biochem. 1990 Jun 20;190(2):233-48. doi: 10.1111/j.1432-1033.1990.tb15568.x. Eur J Biochem. 1990. PMID: 2194799 Review.
Cited by
-
A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein.J Virol. 2002 Oct;76(20):10245-55. doi: 10.1128/jvi.76.20.10245-10255.2002. J Virol. 2002. PMID: 12239300 Free PMC article.
-
Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal.J Virol. 2005 Jan;79(1):132-9. doi: 10.1128/JVI.79.1.132-139.2005. J Virol. 2005. PMID: 15596809 Free PMC article.
-
Bacillus subtilis mutants defective in bacteriophage phi 29 head assembly.J Bacteriol. 1993 Apr;175(8):2357-62. doi: 10.1128/jb.175.8.2357-2362.1993. J Bacteriol. 1993. PMID: 8096839 Free PMC article.
-
Structure and Assembly of TP901-1 Virion Unveiled by Mutagenesis.PLoS One. 2015 Jul 6;10(7):e0131676. doi: 10.1371/journal.pone.0131676. eCollection 2015. PLoS One. 2015. PMID: 26147978 Free PMC article.
-
In vitro incorporation of the phage Phi29 connector complex.Virology. 2009 Nov 10;394(1):149-53. doi: 10.1016/j.virol.2009.08.016. Epub 2009 Sep 9. Virology. 2009. PMID: 19744688 Free PMC article.
References
-
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature (London) 1984;308:32–36. - PubMed
-
- Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophages. Annu Rev Microbiol. 1985;39:109–130. - PubMed
-
- Bazinet C, King J. Initiation of p22 procapsid assembly in vivo. J Mol Biol. 1988;202:77–86. - PubMed
-
- Berget P. Pathways in viral morphogenesis. In: Casjens S, editor. Virus Structure and Assembly. Jones & Bartlett; Boston: 1985. pp. 149–168.
-
- Black LW. DNA packaging in dsDNA bacteriophagse. In: Calendar R, editor. The Bacteriophages. Vol. 2. Plenum; New York: 1988. pp. 321–373.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
