In vitro and in vivo analysis of B-Myb in basal-like breast cancer
- PMID: 19043454
- PMCID: PMC2636852
- DOI: 10.1038/onc.2008.430
In vitro and in vivo analysis of B-Myb in basal-like breast cancer
Abstract
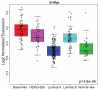
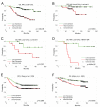
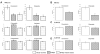
A defining feature of basal-like breast cancer, a breast cancer subtype with poor clinical prognosis, is the high expression of 'proliferation signature' genes. We identified B-Myb, a MYB family transcription factor that is often amplified and overexpressed in many tumor types, as being highly expressed in the proliferation signature. However, the roles of B-Myb in disease progression, and its mammary-specific transcriptional targets, are poorly understood. Here, we showed that B-Myb expression is a significant predictor of survival and pathological complete response to neoadjuvant chemotherapy in breast cancer patients. We also identified a significant association between the G/G genotype of a nonsynonymous B-Myb germline variant (rs2070235, S427G) and an increased risk of basal-like breast cancer [OR 2.0, 95% CI (1.1-3.8)]. In immortalized, human mammary epithelial cell lines, but not in basal-like tumor lines, cells ectopically expressing wild-type B-Myb or the S427G variant showed increased sensitivity to two DNA topoisomerase IIalpha inhibitors, but not to other chemotherapeutics. In addition, microarray analyses identified many G2/M genes as being induced in B-Myb overexpressing cells. These results confirm that B-Myb is involved in cell cycle control, and that its dysregulation may contribute to increased sensitivity to a specific class of chemotherapeutic agents. These data provide insight into the influence of B-Myb in human breast cancer, which is of potential clinical importance for determining disease risk and for guiding treatment.
Figures


Similar articles
-
ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2.Cancer Res. 2010 Nov 15;70(22):9402-12. doi: 10.1158/0008-5472.CAN-10-1199. Epub 2010 Sep 23. Cancer Res. 2010. PMID: 20864510 Free PMC article.
-
Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B-MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy.Oncotarget. 2017 Feb 14;8(7):11160-11172. doi: 10.18632/oncotarget.14466. Oncotarget. 2017. PMID: 28061449 Free PMC article.
-
B-myb is a gene implicated in cell cycle and proliferation of breast cancer.Int J Clin Exp Pathol. 2014 Aug 15;7(9):5819-27. eCollection 2014. Int J Clin Exp Pathol. 2014. Retraction in: Int J Clin Exp Pathol. 2016 Feb 01;9(2):2754. PMID: 25337223 Free PMC article. Retracted.
-
B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments.J Cell Physiol. 1999 Jun;179(3):245-50. doi: 10.1002/(SICI)1097-4652(199906)179:3<245::AID-JCP1>3.0.CO;2-H. J Cell Physiol. 1999. PMID: 10228942 Review.
-
HER-2/neu and topoisomerase IIalpha in breast cancer.Breast Cancer Res Treat. 2003 Apr;78(3):299-311. doi: 10.1023/a:1023077507295. Breast Cancer Res Treat. 2003. PMID: 12755489 Review.
Cited by
-
Differential network analysis applied to preoperative breast cancer chemotherapy response.PLoS One. 2013 Dec 9;8(12):e81784. doi: 10.1371/journal.pone.0081784. eCollection 2013. PLoS One. 2013. PMID: 24349128 Free PMC article.
-
Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes.Breast Cancer Res Treat. 2011 Sep;129(2):593-606. doi: 10.1007/s10549-011-1517-z. Epub 2011 Apr 23. Breast Cancer Res Treat. 2011. PMID: 21516303 Free PMC article.
-
The Proliferative and Apoptotic Landscape of Basal-like Breast Cancer.Int J Mol Sci. 2019 Feb 4;20(3):667. doi: 10.3390/ijms20030667. Int J Mol Sci. 2019. PMID: 30720718 Free PMC article. Review.
-
RAS, cellular senescence and transformation: the BRCA1 DNA repair pathway at the crossroads.Small GTPases. 2012 Jul-Sep;3(3):163-7. doi: 10.4161/sgtp.19884. Epub 2012 Jul 1. Small GTPases. 2012. PMID: 22751483 Free PMC article.
-
Novel Molecular Markers for Breast Cancer.Biomark Cancer. 2016 Mar 13;8:25-42. doi: 10.4137/BIC.S38394. eCollection 2016. Biomark Cancer. 2016. PMID: 26997872 Free PMC article. Review.
References
-
- Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes and Cancer. 2006;45:1033–1040. - PubMed
-
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The Triple Negative Paradox : Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin Cancer Res. 2007;13:2329–2334. - PubMed
-
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. - PubMed
-
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

