Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation
- PMID: 19035365
- PMCID: PMC3081710
- DOI: 10.1002/dvdy.21779
Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation
Abstract
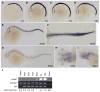
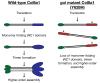
Several zebrafish mutants identified in large-scale forward genetic screens exhibit notochord distortion. We now report the cloning and further characterization of one such mutant, gulliver(m208) (gul(m208)). The notochord defect in gul(m208) mutants is exacerbated under conditions of copper depletion or lysyl oxidase cuproenzyme inhibition that are without a notochord effect on wild-type embryos. The gul(m208) phenotype results from a missense mutation in the gene encoding Col8a1, a lysyl oxidase substrate, and morpholino knockdown of col8a1 recapitulates the notochord distortion observed in gul(m208) mutants. Of interest, the amino acid mutated in gul(m208) Col8a1 is highly conserved, and the equivalent substitution in a closely related human protein, COL10A1, causes Schmid metaphyseal chondrodysplasia. Taken together, the data identify a new protein essential for notochord morphogenesis, extend our understanding of gene-nutrient interactions in early development, and suggest that human mutations in COL8A1 may cause structural birth defects.
(c) 2008 Wiley-Liss, Inc.
Figures



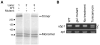

Similar articles
-
Essential role for fibrillin-2 in zebrafish notochord and vascular morphogenesis.Dev Dyn. 2008 Oct;237(10):2844-61. doi: 10.1002/dvdy.21705. Dev Dyn. 2008. PMID: 18816837 Free PMC article.
-
Essential role of lysyl oxidases in notochord development.Dev Biol. 2007 Jul 15;307(2):202-13. doi: 10.1016/j.ydbio.2007.04.029. Epub 2007 May 1. Dev Biol. 2007. PMID: 17543297 Free PMC article.
-
Craniofacial cartilage morphogenesis requires zebrafish col11a1 activity.Matrix Biol. 2009 Oct;28(8):490-502. doi: 10.1016/j.matbio.2009.07.004. Epub 2009 Jul 26. Matrix Biol. 2009. PMID: 19638309
-
Chemical genetics suggests a critical role for lysyl oxidase in zebrafish notochord morphogenesis.Mol Biosyst. 2007 Jan;3(1):51-9. doi: 10.1039/b613673g. Epub 2006 Nov 14. Mol Biosyst. 2007. PMID: 17216056 Free PMC article.
-
Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations.Dev Biol. 2014 Feb 1;386(1):72-85. doi: 10.1016/j.ydbio.2013.11.028. Epub 2013 Dec 11. Dev Biol. 2014. PMID: 24333517 Free PMC article.
Cited by
-
Collagen VIII in vascular diseases.Matrix Biol. 2024 Nov;133:64-76. doi: 10.1016/j.matbio.2024.08.006. Epub 2024 Aug 16. Matrix Biol. 2024. PMID: 39154854 Free PMC article. Review.
-
Coordinated activation of the secretory pathway during notochord formation in the Xenopus embryo.Development. 2009 Nov;136(21):3543-8. doi: 10.1242/dev.036715. Epub 2009 Sep 30. Development. 2009. PMID: 19793890 Free PMC article.
-
A novel zinc finger protein 219-like (ZNF219L) is involved in the regulation of collagen type 2 alpha 1a (col2a1a) gene expression in zebrafish notochord.Int J Biol Sci. 2013 Sep 5;9(9):872-86. doi: 10.7150/ijbs.7126. eCollection 2013. Int J Biol Sci. 2013. PMID: 24155663 Free PMC article.
-
Formation, function, and exhaustion of notochordal cytoplasmic vacuoles within intervertebral disc: current understanding and speculation.Oncotarget. 2017 May 23;8(34):57800-57812. doi: 10.18632/oncotarget.18101. eCollection 2017 Aug 22. Oncotarget. 2017. PMID: 28915712 Free PMC article. Review.
-
A dynamic history of gene duplications and losses characterizes the evolution of the SPARC family in eumetazoans.Proc Biol Sci. 2013 Feb 27;280(1757):20122963. doi: 10.1098/rspb.2012.2963. Print 2013 Apr 22. Proc Biol Sci. 2013. PMID: 23446527 Free PMC article.
References
-
- Bateman JF, Freddi S, McNeil R, Thompson E, Hermanns P, Savarirayan R, Lamande SR. Identification of four novel COL10A1 missense mutations in schmid metaphyseal chondrodysplasia: further evidence that collagen X NC1 mutations impair trimer assembly. Hum Mutat. 2004;23:396. - PubMed
-
- Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet. 2003;12:217–225. - PubMed
-
- Bateman JF, Wilson R, Freddi S, Lamande SR, Savarirayan R. Mutations of COL10A1 in Schmid metaphyseal chondrodysplasia. Hum Mutat. 2005;25:525–534. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

