Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity
- PMID: 19030232
- PMCID: PMC2583956
- DOI: 10.1371/journal.pone.0003802
Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity
Abstract
Background: Identification of genes that modulate longevity is a major focus of aging-related research and an area of intense public interest. In addition to facilitating an improved understanding of the basic mechanisms of aging, such genes represent potential targets for therapeutic intervention in multiple age-associated diseases, including cancer, heart disease, diabetes, and neurodegenerative disorders. To date, however, targeted efforts at identifying longevity-associated genes have been limited by a lack of predictive power, and useful algorithms for candidate gene-identification have also been lacking.
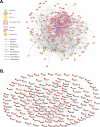
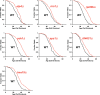
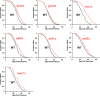
Methodology/principal findings: We have utilized a shortest-path network analysis to identify novel genes that modulate longevity in Saccharomyces cerevisiae. Based on a set of previously reported genes associated with increased life span, we applied a shortest-path network algorithm to a pre-existing protein-protein interaction dataset in order to construct a shortest-path longevity network. To validate this network, the replicative aging potential of 88 single-gene deletion strains corresponding to predicted components of the shortest-path longevity network was determined. Here we report that the single-gene deletion strains identified by our shortest-path longevity analysis are significantly enriched for mutations conferring either increased or decreased replicative life span, relative to a randomly selected set of 564 single-gene deletion strains or to the current data set available for the entire haploid deletion collection. Further, we report the identification of previously unknown longevity genes, several of which function in a conserved longevity pathway believed to mediate life span extension in response to dietary restriction.
Conclusions/significance: This work demonstrates that shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity and represents the first application of network analysis of aging to be extensively validated in a biological system. The novel longevity genes identified in this study are likely to yield further insight into the molecular mechanisms of aging and age-associated disease.
Conflict of interest statement
Figures

Similar articles
-
Deciphering the effects of gene deletion on yeast longevity using network and machine learning approaches.Biochimie. 2012 Apr;94(4):1017-25. doi: 10.1016/j.biochi.2011.12.024. Epub 2012 Jan 8. Biochimie. 2012. PMID: 22239951
-
A genomic analysis of chronological longevity factors in budding yeast.Cell Cycle. 2011 May 1;10(9):1385-96. doi: 10.4161/cc.10.9.15464. Epub 2011 May 1. Cell Cycle. 2011. PMID: 21447998 Free PMC article.
-
Quantitative evidence for conserved longevity pathways between divergent eukaryotic species.Genome Res. 2008 Apr;18(4):564-70. doi: 10.1101/gr.074724.107. Epub 2008 Mar 13. Genome Res. 2008. PMID: 18340043 Free PMC article.
-
Selection for maximum longevity in mice.Exp Gerontol. 1997 Jan-Apr;32(1-2):65-78. doi: 10.1016/s0531-5565(96)00034-4. Exp Gerontol. 1997. PMID: 9088903 Review.
-
Functional genomics of dietary restriction and longevity in yeast.Mech Ageing Dev. 2019 Apr;179:36-43. doi: 10.1016/j.mad.2019.02.003. Epub 2019 Feb 18. Mech Ageing Dev. 2019. PMID: 30790575 Review.
Cited by
-
Cryptococcus neoformans constitutes an ideal model organism to unravel the contribution of cellular aging to the virulence of chronic infections.Curr Opin Microbiol. 2013 Aug;16(4):391-7. doi: 10.1016/j.mib.2013.03.011. Epub 2013 Apr 27. Curr Opin Microbiol. 2013. PMID: 23631868 Free PMC article. Review.
-
Whi2 signals low leucine availability to halt yeast growth and cell death.FEMS Yeast Res. 2018 Dec 1;18(8):foy095. doi: 10.1093/femsyr/foy095. FEMS Yeast Res. 2018. PMID: 30165592 Free PMC article. Review.
-
Distinctive topology of age-associated epigenetic drift in the human interactome.Proc Natl Acad Sci U S A. 2013 Aug 27;110(35):14138-43. doi: 10.1073/pnas.1307242110. Epub 2013 Aug 12. Proc Natl Acad Sci U S A. 2013. PMID: 23940324 Free PMC article.
-
Cytoplasmic and Mitochondrial NADPH-Coupled Redox Systems in the Regulation of Aging.Nutrients. 2019 Feb 27;11(3):504. doi: 10.3390/nu11030504. Nutrients. 2019. PMID: 30818813 Free PMC article. Review.
-
Chapter 5: Network biology approach to complex diseases.PLoS Comput Biol. 2012;8(12):e1002820. doi: 10.1371/journal.pcbi.1002820. Epub 2012 Dec 27. PLoS Comput Biol. 2012. PMID: 23300411 Free PMC article.
References
-
- Barabasi A-L, Oltvai ZN. Network Biology: Understanding the cell's functional organization. Nature Reviews: Genetics. 2004;5:101–113. - PubMed
-
- Bonchev D. Complexity analysis of yeast proteome network. Chem Biodivers. 2004;1:312–326. - PubMed
-
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and Centrality in Protein Networks. Nature. 2001;411:41–42. - PubMed
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

