The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats
- PMID: 19017650
- PMCID: PMC3471988
- DOI: 10.1074/jbc.M805113200
The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats
Abstract
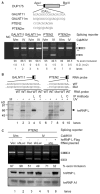
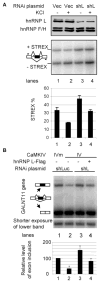
The regulation of gene expression through alternative pre-mRNA splicing is common in metazoans and is often controlled by intracellular signaling pathways that are important in cell physiology. We have shown that the alternative splicing of a number of genes is controlled by membrane depolarization and Ca2+/calmodulin-dependent protein kinase IV (CaMKIV) through CaMKIV-responsive RNA elements (CaRRE1 and CaRRE2); however, the trans-acting factors remain unknown. Here we show that the heterogeneous nuclear ribonucleoprotein (hnRNP) L is a CaRRE1 binding factor in nuclear extracts. An hnRNP L high affinity CA (cytidine-adenosine) repeat element is sufficient to mediate CaMKIV and hnRNP L repression of splicing in a location (3'-splice site proximity)-dependent way. Depletion of hnRNP L by RNA interference followed by rescue with coexpressed exogenous hnRNP L demonstrates that hnRNP L mediates the CaMKIV-regulated splicing through CA repeats in heterologous contexts. Depletion of hnRNP L also led to increased inclusion of the stress axis-regulated exon and a CA repeat-harboring exon under depolarization or with activated CaMKIV. Moreover, hnRNP L binding to CaRRE1 was increased by CaMKIV and, conversely, was reduced by pretreatments with protein phosphatases. Therefore, hnRNP L is an essential component of CaMKIV-regulated alternative splicing through CA repeats, with its phosphorylation likely playing a critical role.
Figures



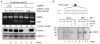

Similar articles
-
A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates depolarization-regulated alternative splicing of potassium channels.J Biol Chem. 2012 Jun 29;287(27):22709-16. doi: 10.1074/jbc.M112.357343. Epub 2012 May 8. J Biol Chem. 2012. PMID: 22570490 Free PMC article.
-
In vivo selection of kinase-responsive RNA elements controlling alternative splicing.J Biol Chem. 2009 Jun 12;284(24):16191-16201. doi: 10.1074/jbc.M900393200. Epub 2009 Apr 22. J Biol Chem. 2009. PMID: 19386606 Free PMC article.
-
Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M.J Biol Chem. 2011 May 6;286(18):16039-51. doi: 10.1074/jbc.M110.204057. Epub 2011 Mar 11. J Biol Chem. 2011. PMID: 21398516 Free PMC article.
-
Differential evolution of signal-responsive RNA elements and upstream factors that control alternative splicing.Cell Mol Life Sci. 2014 Nov;71(22):4347-60. doi: 10.1007/s00018-014-1688-y. Epub 2014 Jul 27. Cell Mol Life Sci. 2014. PMID: 25064062 Free PMC article. Review.
-
Alternative splicing: multiple control mechanisms and involvement in human disease.Trends Genet. 2002 Apr;18(4):186-93. doi: 10.1016/s0168-9525(01)02626-9. Trends Genet. 2002. PMID: 11932019 Review.
Cited by
-
A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability.Am J Hum Genet. 2012 Oct 5;91(4):694-702. doi: 10.1016/j.ajhg.2012.08.011. Epub 2012 Sep 20. Am J Hum Genet. 2012. PMID: 23000143 Free PMC article.
-
Widespread Separation of the Polypyrimidine Tract From 3' AG by G Tracts in Association With Alternative Exons in Metazoa and Plants.Front Genet. 2019 Jan 14;9:741. doi: 10.3389/fgene.2018.00741. eCollection 2018. Front Genet. 2019. PMID: 30693020 Free PMC article.
-
Common variants in genes of the postsynaptic FMRP signalling pathway are risk factors for autism spectrum disorders.Hum Genet. 2014 Jun;133(6):781-92. doi: 10.1007/s00439-013-1416-y. Epub 2014 Jan 19. Hum Genet. 2014. PMID: 24442360
-
Multilevel Differential Control of Hormone Gene Expression Programs by hnRNP L and LL in Pituitary Cells.Mol Cell Biol. 2018 May 29;38(12):e00651-17. doi: 10.1128/MCB.00651-17. Print 2018 Jun 15. Mol Cell Biol. 2018. PMID: 29610151 Free PMC article.
-
Mechanism of alternative splicing and its regulation.Biomed Rep. 2015 Mar;3(2):152-158. doi: 10.3892/br.2014.407. Epub 2014 Dec 17. Biomed Rep. 2015. PMID: 25798239 Free PMC article.
References
-
- Black DL. Annu Rev Biochem. 2003;72:291–336. - PubMed
-
- Black DL. Cell. 2000;103:367–370. - PubMed
-
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Science. 2003;302:2141–2144. - PubMed
-
- Graveley BR. Trends Genet. 2001;17:100–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

