Structural insights into phenylethanolamines high-affinity binding site in NR2B from binding and molecular modeling studies
- PMID: 19017396
- PMCID: PMC2603005
- DOI: 10.1186/1756-6606-1-16
Structural insights into phenylethanolamines high-affinity binding site in NR2B from binding and molecular modeling studies
Abstract
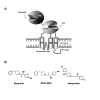
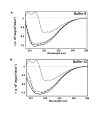
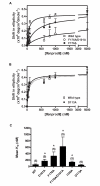
Background: Phenylethanolamines selectively bind to NR2B subunit-containing N-methyl-D-aspartate-subtype of ionotropic glutamate receptors and negatively modulate receptor activity. To investigate the structural and functional properties of the ifenprodil binding domain on the NR2B protein, we have purified a soluble recombinant rat NR2B protein fragment comprising the first ~400 amino acid amino-terminal domain (ATD2B) expressed in E. coli. Spectral measurements on refolded ATD2B protein demonstrated specific binding to ifenprodil. We have used site-directed mutagenesis, circular dichroism spectroscopy and molecular modeling to obtain structural information on the interactions between critical amino acid residues and ifenprodil of our soluble refolded ATD2B proteins. Ligand-induced changes in protein structure were inferred from changes in the circular dichroism spectrum, and the concentration dependence of these changes was used to determine binding constants for ifenprodil and its analogues.
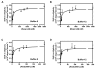
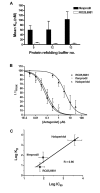
Results: Ligand binding of ifenprodil, RO25,6981 and haloperidol on soluble recombinant ATD2B determined from circular dichroism spectroscopy yielded low-to-high micromolar equilibrium constants which concurred with functional IC₅₀ measurement determined in heterologously expressed NR1/NR2B receptors in Xenopus oocytes. Amino acid residue substitutions of Asp101, Ile150 and Phe176 with alanine residue within the ATD2B protein altered the recombinant protein dissociation constants for ifenprodil, mirroring the pattern of their functional phenotypes. Molecular modeling of ATD2B as a clam-shell-like structure places these critical residues near a putative ligand binding site.
Conclusion: We report for the first time biochemical measurements show that the functional measurements actually reflect binding to the ATD of NR2B subunit. Insights gained from this study help advance the theory that ifenprodil is a ligand for the ATD of NR2B subunit.
Figures

Similar articles
-
Expression and characterization of soluble amino-terminal domain of NR2B subunit of N-methyl-D-aspartate receptor.Protein Sci. 2005 Sep;14(9):2275-83. doi: 10.1110/ps.051509905. Protein Sci. 2005. PMID: 16131656 Free PMC article.
-
Improving solubility of NR2B amino-terminal domain of N-methyl-d-aspartate receptor expressed in Escherichia coli.Biochem Biophys Res Commun. 2007 Oct 12;362(1):69-74. doi: 10.1016/j.bbrc.2007.07.164. Epub 2007 Aug 8. Biochem Biophys Res Commun. 2007. PMID: 17706601
-
Binding of spermine and ifenprodil to a purified, soluble regulatory domain of the N-methyl-D-aspartate receptor.J Neurochem. 2008 Dec;107(6):1566-77. doi: 10.1111/j.1471-4159.2008.05729.x. Epub 2008 Nov 5. J Neurochem. 2008. PMID: 19014388 Free PMC article.
-
Ifenprodil, a novel NMDA receptor antagonist: site and mechanism of action.Curr Drug Targets. 2001 Sep;2(3):285-98. doi: 10.2174/1389450013348489. Curr Drug Targets. 2001. PMID: 11554553 Review.
-
The NMDA receptor NR2B subunit: a valid therapeutic target for multiple CNS pathologies.Curr Med Chem. 2004 Feb;11(3):389-96. doi: 10.2174/0929867043456061. Curr Med Chem. 2004. PMID: 14965239 Review.
Cited by
-
Mapping the high-affinity binding domain of 5-substituted benzimidazoles to the proximal N-terminus of the GluN2B subunit of the NMDA receptor.Br J Pharmacol. 2010 Jan 1;159(2):449-61. doi: 10.1111/j.1476-5381.2009.00549.x. Epub 2010 Jan 15. Br J Pharmacol. 2010. PMID: 20082612 Free PMC article.
-
Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential.Br J Pharmacol. 2009 Aug;157(8):1301-17. doi: 10.1111/j.1476-5381.2009.00304.x. Epub 2009 Jul 8. Br J Pharmacol. 2009. PMID: 19594762 Free PMC article. Review.
-
Control of assembly and function of glutamate receptors by the amino-terminal domain.Mol Pharmacol. 2010 Oct;78(4):535-49. doi: 10.1124/mol.110.067157. Epub 2010 Jul 21. Mol Pharmacol. 2010. PMID: 20660085 Free PMC article. Review.
-
Molecular bases of NMDA receptor subtype-dependent properties.J Physiol. 2015 Jan 1;593(1):83-95. doi: 10.1113/jphysiol.2014.273763. Epub 2014 Sep 9. J Physiol. 2015. PMID: 25556790 Free PMC article. Review.
-
Control of NMDA receptor function by the NR2 subunit amino-terminal domain.J Neurosci. 2009 Sep 30;29(39):12045-58. doi: 10.1523/JNEUROSCI.1365-09.2009. J Neurosci. 2009. PMID: 19793963 Free PMC article.
References
-
- Massey PV, Johnson BE, Moult PR, Auberson YP, Brown MW, Molnar E, Collingridge GL, Bashir ZI. Differential roles of NR2A and NR2B-containing NMDA receptors in cortical long-term potentiation and long-term depression. J Neurosci. 2004;24:7821–7828. doi: 10.1523/JNEUROSCI.1697-04.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

