The substrate recognition domains of the N-end rule pathway
- PMID: 19008229
- PMCID: PMC2615520
- DOI: 10.1074/jbc.M803641200
The substrate recognition domains of the N-end rule pathway
Abstract
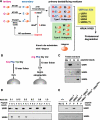
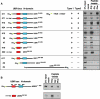
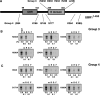
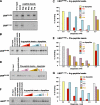
The N-end rule pathway is a ubiquitin-dependent system where E3 ligases called N-recognins, including UBR1 and UBR2, recognize type-1 (basic) and type-2 (bulky hydrophobic) N-terminal residues as part of N-degrons. We have recently reported an E3 family (termed UBR1 through UBR7) characterized by the 70-residue UBR box, among which UBR1, UBR2, UBR4, and UBR5 were captured during affinity-based proteomics with synthetic degrons. Here we characterized substrate binding specificity and recognition domains of UBR proteins. Pull-down assays with recombinant UBR proteins suggest that 570-kDa UBR4 and 300-kDa UBR5 bind N-degron, whereas UBR3, UBR6, and UBR7 do not. Binding assays with 24 UBR1 deletion mutants and 31 site-directed UBR1 mutations narrow down the degron-binding activity to a 72-residue UBR box-only fragment that recognizes type-1 but not type-2 residues. A surface plasmon resonance assay shows that the UBR box binds to the type-1 substrate Arg-peptide with Kd of approximately 3.4 microm. Downstream from the UBR box, we identify a second substrate recognition domain, termed the N-domain, required for type-2 substrate recognition. The approximately 80-residue N-domain shows structural and functional similarity to 106-residue Escherichia coli ClpS, a bacterial N-recognin. We propose a model where the 70-residue UBR box functions as a common structural element essential for binding to all known destabilizing N-terminal residues, whereas specific residues localized in the UBR box (for type 1) or the N-domain (for type 2) provide substrate selectivity through interaction with the side group of an N-terminal amino acid. Our work provides new insights into substrate recognition in the N-end rule pathway.
Figures





Similar articles
-
A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons.Mol Cell Biol. 2005 Aug;25(16):7120-36. doi: 10.1128/MCB.25.16.7120-7136.2005. Mol Cell Biol. 2005. PMID: 16055722 Free PMC article.
-
Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems.J Biol Chem. 2007 Jun 22;282(25):18510-18520. doi: 10.1074/jbc.M701894200. Epub 2007 Apr 26. J Biol Chem. 2007. PMID: 17462990
-
Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway.J Biol Chem. 2008 Aug 29;283(35):24011-28. doi: 10.1074/jbc.M802583200. Epub 2008 Jun 19. J Biol Chem. 2008. PMID: 18566452 Free PMC article.
-
Signaling Pathways Regulated by UBR Box-Containing E3 Ligases.Int J Mol Sci. 2021 Aug 3;22(15):8323. doi: 10.3390/ijms22158323. Int J Mol Sci. 2021. PMID: 34361089 Free PMC article. Review.
-
Multivalency-assisted control of intracellular signaling pathways: application for ubiquitin- dependent N-end rule pathway.Chem Biol. 2009 Feb 27;16(2):121-31. doi: 10.1016/j.chembiol.2009.01.012. Chem Biol. 2009. PMID: 19246002 Free PMC article. Review.
Cited by
-
Physiological functions and clinical implications of the N-end rule pathway.Front Med. 2016 Sep;10(3):258-70. doi: 10.1007/s11684-016-0458-7. Epub 2016 Sep 7. Front Med. 2016. PMID: 27492620 Review.
-
Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations.PLoS One. 2009 Nov 13;4(11):e7757. doi: 10.1371/journal.pone.0007757. PLoS One. 2009. PMID: 19915679 Free PMC article.
-
The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development.Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13618-23. doi: 10.1073/pnas.0906404106. Epub 2009 Jul 20. Proc Natl Acad Sci U S A. 2009. PMID: 19620738 Free PMC article.
-
Starting with a degron: N-terminal formyl-methionine of nascent bacterial proteins contributes to their proteolytic control.Microb Cell. 2015 Oct 5;2(10):356-359. doi: 10.15698/mic2015.10.235. Microb Cell. 2015. PMID: 28357262 Free PMC article. No abstract available.
-
Manumycin polyketides act as molecular glues between UBR7 and P53.Nat Chem Biol. 2020 Nov;16(11):1189-1198. doi: 10.1038/s41589-020-0557-2. Epub 2020 Jun 22. Nat Chem Biol. 2020. PMID: 32572277 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

