Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease
- PMID: 18996898
- PMCID: PMC2602775
- DOI: 10.1093/nar/gkn880
Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease
Abstract
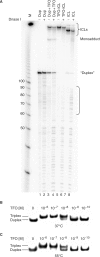
Photoreactive psoralens can form interstrand crosslinks (ICLs) in double-stranded DNA. In eubacteria, the endonuclease UvrABC plays a key role in processing psoralen ICLs. Psoralen-modified triplex-forming oligonucleotides (TFOs) can be used to direct ICLs to specific genomic sites. Previous studies of pyrimidine-rich methoxypsoralen-modified TFOs indicated that the TFO inhibits cleavage by UvrABC. Because different chemistries may alter the processing of TFO-directed ICLs, we investigated the effect of another type of triplex formed by purine-rich TFOs on the processing of 4'-(hydroxymethyl)-4,5',8-trimethylpsoralen (HMT) ICLs by the UvrABC nuclease. Using an HMT-modified TFO to direct ICLs to a specific site, we found that UvrABC made incisions on the purine-rich strand of the duplex approximately 3 bases from the 3'-side and approximately 9 bases from the 5'-side of the ICL, within the TFO-binding region. In contrast to previous reports, the UvrABC nuclease cleaved the TFO-directed psoralen ICL with a greater efficiency than that of the psoralen ICL alone. Furthermore, the TFO was dissociated from its duplex binding site by UvrA and UvrB. As mutagenesis by TFO-directed ICLs requires nucleotide excision repair, the efficient processing of these lesions supports the use of triplex technology to direct DNA damage for genome modification.
Figures


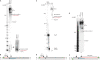


Similar articles
-
Targeting and processing of site-specific DNA interstrand crosslinks.Environ Mol Mutagen. 2010 Jul;51(6):527-39. doi: 10.1002/em.20557. Environ Mol Mutagen. 2010. PMID: 20196133 Free PMC article. Review.
-
Tools to Study the Role of Architectural Protein HMGB1 in the Processing of Helix Distorting, Site-specific DNA Interstrand Crosslinks.J Vis Exp. 2016 Nov 10;(117):54678. doi: 10.3791/54678. J Vis Exp. 2016. PMID: 27911399 Free PMC article.
-
Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks.Nucleic Acids Res. 2009 Jul;37(13):4420-9. doi: 10.1093/nar/gkp399. Epub 2009 May 25. Nucleic Acids Res. 2009. PMID: 19468048 Free PMC article.
-
The repair of psoralen monoadducts by the Escherichia coli UvrABC endonuclease.Nucleic Acids Res. 1987 Jun 25;15(12):4957-71. doi: 10.1093/nar/15.12.4957. Nucleic Acids Res. 1987. PMID: 3299260 Free PMC article.
-
Triplex-forming oligonucleotides as potential tools for modulation of gene expression.Curr Med Chem Anticancer Agents. 2005 Jul;5(4):319-26. doi: 10.2174/1568011054222300. Curr Med Chem Anticancer Agents. 2005. PMID: 16101484 Review.
Cited by
-
Mammalian nucleotide excision repair proteins and interstrand crosslink repair.Environ Mol Mutagen. 2010 Jul;51(6):520-6. doi: 10.1002/em.20569. Environ Mol Mutagen. 2010. PMID: 20658645 Free PMC article. Review.
-
Targeting and processing of site-specific DNA interstrand crosslinks.Environ Mol Mutagen. 2010 Jul;51(6):527-39. doi: 10.1002/em.20557. Environ Mol Mutagen. 2010. PMID: 20196133 Free PMC article. Review.
-
Targeted gene conversion induced by triplex-directed psoralen interstrand crosslinks in mammalian cells.Nucleic Acids Res. 2009 Oct;37(19):6378-88. doi: 10.1093/nar/gkp678. Epub 2009 Sep 2. Nucleic Acids Res. 2009. PMID: 19726585 Free PMC article.
-
UvrA expression of Lactococcus lactis NZ9000 improve multiple stresses tolerance and fermentation of lactic acid against salt stress.J Food Sci Technol. 2017 Mar;54(3):639-649. doi: 10.1007/s13197-017-2493-z. Epub 2017 Feb 22. J Food Sci Technol. 2017. PMID: 28298677 Free PMC article.
-
DNA interstrand crosslink repair in mammalian cells: step by step.Crit Rev Biochem Mol Biol. 2010 Feb;45(1):23-49. doi: 10.3109/10409230903501819. Crit Rev Biochem Mol Biol. 2010. PMID: 20039786 Free PMC article. Review.
References
-
- Stern RS. Psoralen and ultraviolet a light therapy for psoriasis. N Engl J. Med. 2007;357:682–690. - PubMed
-
- Song PS, Tapley K.J., Jr Photochemistry and photobiology of psoralens. Photochem. Photobiol. 1979;29:1177–1197. - PubMed
-
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1154–1193. - PubMed
-
- Shi Y-B, Hearst JE. Wavelength dependence for the photoreactions of DNA-psoralen monoadducts. 2. Photo-cross-linking of monoadducts. Biochemistry. 1987;26:3792–3798. - PubMed
-
- Van Houten B, Gamper H, Hearst JE, Sancar A. Construction of DNA substrates modified with psoralen at a unique site and study of the action mechanism of ABC excinuclease on these uniformly modified substrates. J. Biol. Chem. 1986;261:14135–14141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

