Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha
- PMID: 18976920
- PMCID: PMC3556578
- DOI: 10.1016/j.tim.2008.08.013
Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha
Abstract
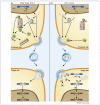
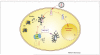
Pathogenic viral infections have exerted selection pressure on their hosts to evolve cellular antiviral inhibitors referred to as restriction factors. Examples of such molecules are APOBEC3G, APOBEC3F and TRIM5alpha. APOBEC3G and APOBEC3F are cytidine deaminases that are able to strongly inhibit retroviral replication by at least two mechanisms. They are counteracted by the lentiviral Vif protein. TRIM5alpha binds to sensitive, incoming retroviruses via its C-terminal PRY/SPRY domain and rapidly recruits them to the proteasome before significant viral DNA synthesis can occur. Both of these proteins robustly block retroviral replication in a species-specific way. It remains an open but important question as to whether innate restriction factors such as these can be harnessed to inhibit HIV-1 replication in humans.
Figures

Similar articles
-
Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation.J Virol. 2014 May;88(10):5881-7. doi: 10.1128/JVI.00062-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623435 Free PMC article.
-
HIV Restriction Factors and Mechanisms of Evasion.Cold Spring Harb Perspect Med. 2012 May;2(5):a006940. doi: 10.1101/cshperspect.a006940. Cold Spring Harb Perspect Med. 2012. PMID: 22553496 Free PMC article. Review.
-
APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G.J Biol Chem. 2007 Jan 26;282(4):2587-95. doi: 10.1074/jbc.M607298200. Epub 2006 Nov 22. J Biol Chem. 2007. PMID: 17121840
-
Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities.Nucleic Acids Res. 2005 Apr 4;33(6):1913-23. doi: 10.1093/nar/gki343. Print 2005. Nucleic Acids Res. 2005. PMID: 15809227 Free PMC article.
-
[Mechanisms for inhibition of retrovirus replication by APOBEC3 family].Uirusu. 2011 Jun;61(1):67-72. doi: 10.2222/jsv.61.67. Uirusu. 2011. PMID: 21972557 Review. Japanese.
Cited by
-
Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions.Viruses. 2021 Mar 17;13(3):497. doi: 10.3390/v13030497. Viruses. 2021. PMID: 33802945 Free PMC article. Review.
-
Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion.PLoS Pathog. 2009 May;5(5):e1000443. doi: 10.1371/journal.ppat.1000443. Epub 2009 May 22. PLoS Pathog. 2009. PMID: 19461879 Free PMC article.
-
A novel mosquito ubiquitin targets viral envelope protein for degradation and reduces virion production during dengue virus infection.Biochim Biophys Acta. 2016 Sep;1860(9):1898-909. doi: 10.1016/j.bbagen.2016.05.033. Epub 2016 May 27. Biochim Biophys Acta. 2016. PMID: 27241849 Free PMC article.
-
Homology-based identification of capsid determinants that protect HIV1 from human TRIM5α restriction.J Biol Chem. 2011 Mar 11;286(10):8128-8140. doi: 10.1074/jbc.M110.187609. Epub 2010 Dec 17. J Biol Chem. 2011. PMID: 21169362 Free PMC article.
-
Involvement of endogenous retroviruses in prion diseases.Pathogens. 2013 Aug 12;2(3):533-43. doi: 10.3390/pathogens2030533. Pathogens. 2013. PMID: 25437206 Free PMC article. Review.
References
-
- Kaiser J. AIDS research: review of vaccine failure prompts a return to basics. Science. 2008;320:30–31. - PubMed
-
- Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. - PubMed
-
- Sheehy AM, et al. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. - PubMed
-
- Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

