In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites
- PMID: 18950525
- PMCID: PMC2613160
- DOI: 10.1186/1471-2105-9-454
In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites
Abstract
Background: Secondary metabolites biosynthesized by polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) family of enzymes constitute several classes of therapeutically important natural products like erythromycin, rapamycin, cyclosporine etc. In view of their relevance for natural product based drug discovery, identification of novel secondary metabolite natural products by genome mining has been an area of active research. A number of different tailoring enzymes catalyze a variety of chemical modifications to the polyketide or nonribosomal peptide backbone of these secondary metabolites to enhance their structural diversity. Therefore, development of powerful bioinformatics methods for identification of these tailoring enzymes and assignment of their substrate specificity is crucial for deciphering novel secondary metabolites by genome mining.
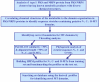
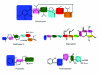
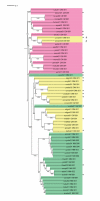
Results: In this work, we have carried out a comprehensive bioinformatics analysis of methyltransferase (MT) domains present in multi functional type I PKS and NRPS proteins encoded by PKS/NRPS gene clusters having known secondary metabolite products. Based on the results of this analysis, we have developed a novel knowledge based computational approach for detecting MT domains present in PKS and NRPS megasynthases, delineating their correct boundaries and classifying them as N-MT, C-MT and O-MT using profile HMMs. Analysis of proteins in nr database of NCBI using these class specific profiles has revealed several interesting examples, namely, C-MT domains in NRPS modules, N-MT domains with significant homology to C-MT proteins, and presence of NRPS/PKS MTs in association with other catalytic domains. Our analysis of the chemical structures of the secondary metabolites and their site of methylation suggested that a possible evolutionary basis for the presence of a novel class of N-MT domains with significant homology to C-MT proteins could be the close resemblance of the chemical structures of the acceptor substrates, as in the case of pyochelin and yersiniabactin. These two classes of MTs recognize similar acceptor substrates, but transfer methyl groups to N and C positions on these substrates.
Conclusion: We have developed a novel knowledge based computational approach for identifying MT domains present in type I PKS and NRPS multifunctional enzymes and predicting their site of methylation. Analysis of nr database using this approach has revealed presence of several novel MT domains. Our analysis has also given interesting insight into the evolutionary basis of the novel substrate specificities of these MT proteins.
Figures

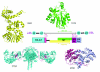




Similar articles
-
Biosynthesis of hybrid peptide-polyketide natural products.Curr Opin Drug Discov Devel. 2001 Mar;4(2):215-28. Curr Opin Drug Discov Devel. 2001. PMID: 11378961 Review.
-
Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics.Biotechnol Lett. 2012 Aug;34(8):1393-403. doi: 10.1007/s10529-012-0919-2. Epub 2012 Apr 6. Biotechnol Lett. 2012. PMID: 22481301 Review.
-
Sequencing and modular analysis of the hybrid non-ribosomal peptide synthase - polyketide synthase gene cluster from the marine sponge Hymeniacidon perleve-associated bacterium Pseudoalteromonas sp. strain NJ631.Can J Microbiol. 2009 Mar;55(3):219-27. doi: 10.1139/w08-125. Can J Microbiol. 2009. PMID: 19370064
-
Chapter 10 using phosphopantetheinyl transferases for enzyme posttranslational activation, site specific protein labeling and identification of natural product biosynthetic gene clusters from bacterial genomes.Methods Enzymol. 2009;458:255-75. doi: 10.1016/S0076-6879(09)04810-1. Methods Enzymol. 2009. PMID: 19374986
-
Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases.Chembiochem. 2006 Aug;7(8):1206-20. doi: 10.1002/cbic.200600075. Chembiochem. 2006. PMID: 16835859
Cited by
-
Combining Mass Spectrometric Metabolic Profiling with Genomic Analysis: A Powerful Approach for Discovering Natural Products from Cyanobacteria.J Nat Prod. 2015 Jul 24;78(7):1671-82. doi: 10.1021/acs.jnatprod.5b00301. Epub 2015 Jul 7. J Nat Prod. 2015. PMID: 26149623 Free PMC article.
-
Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM).Nucleic Acids Res. 2015 Nov 16;43(20):9645-62. doi: 10.1093/nar/gkv1012. Epub 2015 Oct 5. Nucleic Acids Res. 2015. PMID: 26442528 Free PMC article.
-
Bioinformatic Identification of Novel Methyltransferases.Epigenomics. 2009 Oct 1;1(1):163-175. doi: 10.2217/epi.09.3. Epigenomics. 2009. PMID: 20582239 Free PMC article.
-
Initiating polyketide biosynthesis by on-line methyl esterification.Nat Commun. 2021 Jul 23;12(1):4499. doi: 10.1038/s41467-021-24846-7. Nat Commun. 2021. PMID: 34301953 Free PMC article.
-
In silico identification of AMPylating enzymes and study of their divergent evolution.Sci Rep. 2015 Jun 3;5:10804. doi: 10.1038/srep10804. Sci Rep. 2015. PMID: 26039278 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

