The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G)
- PMID: 18948280
- PMCID: PMC2588517
- DOI: 10.1093/nar/gkn647
The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G)
Abstract
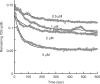
Ribosome recycling factor (RRF), elongation factor G (EF-G) and GTP split 70S ribosomes into subunits. Here, we demonstrated that the splitting was transient and the exhaustion of GTP resulted in re-association of the split subunits into 70S ribosomes unless IF3 (initiation factor 3) was present. However, the splitting was observed with sucrose density gradient centrifugation (SDGC) without IF3 if RRF, EF-G and GTP were present in the SDGC buffer. The splitting of 70S ribosomes causes the decrease of light scattering by ribosomes. Kinetic constants obtained from the light scattering studies are sufficient to account for the splitting of 70S ribosomes by RRF and EF-G/GTP during the lag phase for activation of ribosomes for the log phase. As the amount of 70S ribosomes increased, more RRF, EF-G and GTP were necessary to split 70S ribosomes. In the presence of a physiological amount of polyamines, GTP and factors, even 0.6 microM 70S ribosomes (12 times higher than the 70S ribosomes for routine assay) were split. Spermidine (2 mM) completely inhibited anti-association activity of IF3, and the RRF/EF-G/GTP-dependent splitting of 70S ribosomes.
Figures





Similar articles
-
The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits.RNA. 2005 Aug;11(8):1317-28. doi: 10.1261/rna.2520405. RNA. 2005. PMID: 16043510 Free PMC article.
-
Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting.EMBO J. 2008 Jun 18;27(12):1706-17. doi: 10.1038/emboj.2008.99. Epub 2008 May 22. EMBO J. 2008. PMID: 18497739 Free PMC article.
-
Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G.Mol Cell. 2005 Jun 10;18(6):675-86. doi: 10.1016/j.molcel.2005.05.016. Mol Cell. 2005. PMID: 15949442
-
Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors.Biochimie. 1996;78(11-12):959-69. doi: 10.1016/s0300-9084(97)86718-1. Biochimie. 1996. PMID: 9150873 Review.
-
[Ribosome recycling revisited].Mol Biol (Mosk). 2006 Jul-Aug;40(4):742-50. Mol Biol (Mosk). 2006. PMID: 16913233 Review. Russian.
Cited by
-
Distinct functions of elongation factor G in ribosome recycling and translocation.RNA. 2009 May;15(5):772-80. doi: 10.1261/rna.1592509. Epub 2009 Mar 26. RNA. 2009. PMID: 19324963 Free PMC article.
-
Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.Cell. 2024 Feb 1;187(3):782-796.e23. doi: 10.1016/j.cell.2023.12.027. Epub 2024 Jan 19. Cell. 2024. PMID: 38244547 Free PMC article.
-
The hibernating 100S complex is a target of ribosome-recycling factor and elongation factor G in Staphylococcus aureus.J Biol Chem. 2020 May 1;295(18):6053-6063. doi: 10.1074/jbc.RA119.012307. Epub 2020 Mar 24. J Biol Chem. 2020. PMID: 32209660 Free PMC article.
-
New insights into the enzymatic role of EF-G in ribosome recycling.Nucleic Acids Res. 2015 Dec 2;43(21):10525-33. doi: 10.1093/nar/gkv995. Epub 2015 Oct 1. Nucleic Acids Res. 2015. PMID: 26432831 Free PMC article.
-
Ribosome-dependent ATPase interacts with conserved membrane protein in Escherichia coli to modulate protein synthesis and oxidative phosphorylation.PLoS One. 2011 Apr 27;6(4):e18510. doi: 10.1371/journal.pone.0018510. PLoS One. 2011. PMID: 21556145 Free PMC article.
References
-
- Weaver RF. Molecular biology. 3rd edn. New York: McGraw Hill; 2005.
-
- Frolova L, Goff XL, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. - PubMed
-
- Capecchi MR, Klein HA. Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harb. Symp. Quant. Biol. 1969;34:469–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

