Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii
- PMID: 18931120
- PMCID: PMC2593238
- DOI: 10.1128/JB.00834-08
Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii
Abstract
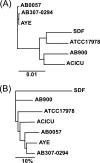
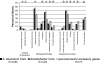
The recent emergence of multidrug resistance (MDR) in Acinetobacter baumannii has raised concern in health care settings worldwide. In order to understand the repertoire of resistance determinants and their organization and origins, we compared the genome sequences of three MDR and three drug-susceptible A. baumannii isolates. The entire MDR phenotype can be explained by the acquisition of discrete resistance determinants distributed throughout the genome. A comparison of closely related MDR and drug-susceptible isolates suggests that drug efflux may be a less significant contributor to resistance to certain classes of antibiotics than inactivation enzymes are. A resistance island with a variable composition of resistance determinants interspersed with transposons, integrons, and other mobile genetic elements is a significant but not universal contributor to the MDR phenotype. Four hundred seventy-five genes are shared among all six clinical isolates but absent from the related environmental species Acinetobacter baylyi ADP1. These genes are enriched for transcription factors and transporters and suggest physiological features of A. baumannii that are related to adaptation for growth in association with humans.
Figures

Similar articles
-
Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance.J Antimicrob Chemother. 2012 Dec;67(12):2825-32. doi: 10.1093/jac/dks327. Epub 2012 Sep 5. J Antimicrob Chemother. 2012. PMID: 22952140
-
Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants.BMC Genomics. 2014 Dec 22;15(1):1163. doi: 10.1186/1471-2164-15-1163. BMC Genomics. 2014. PMID: 25534766 Free PMC article.
-
Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage.Int J Antimicrob Agents. 2015 Nov;46(5):526-31. doi: 10.1016/j.ijantimicag.2015.07.012. Epub 2015 Aug 28. Int J Antimicrob Agents. 2015. PMID: 26362951
-
Genetic basis of antibiotic resistance in pathogenic Acinetobacter species.IUBMB Life. 2011 Dec;63(12):1061-7. doi: 10.1002/iub.532. Epub 2011 Oct 12. IUBMB Life. 2011. PMID: 21990280 Review.
-
Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii.Drug Resist Updat. 2012 Aug;15(4):237-47. doi: 10.1016/j.drup.2012.06.001. Epub 2012 Jul 27. Drug Resist Updat. 2012. PMID: 22841809 Review.
Cited by
-
Variations of AbaR4-type resistance islands in Acinetobacter baumannii isolates from South Korea.Antimicrob Agents Chemother. 2012 Aug;56(8):4544-7. doi: 10.1128/AAC.00880-12. Epub 2012 Jun 5. Antimicrob Agents Chemother. 2012. PMID: 22668861 Free PMC article.
-
CRISPR-cas subtype I-Fb in Acinetobacter baumannii: evolution and utilization for strain subtyping.PLoS One. 2015 Feb 23;10(2):e0118205. doi: 10.1371/journal.pone.0118205. eCollection 2015. PLoS One. 2015. PMID: 25706932 Free PMC article.
-
Heme uptake and utilization by hypervirulent Acinetobacter baumannii LAC-4 is dependent on a canonical heme oxygenase (abHemO).Arch Biochem Biophys. 2019 Sep 15;672:108066. doi: 10.1016/j.abb.2019.108066. Epub 2019 Aug 6. Arch Biochem Biophys. 2019. PMID: 31398314 Free PMC article.
-
Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.Biochemistry. 2010 Nov 2;49(43):9292-305. doi: 10.1021/bi101226n. Biochemistry. 2010. PMID: 20853905 Free PMC article.
-
Integrase-Controlled Excision of Metal-Resistance Genomic Islands in Acinetobacter baumannii.Genes (Basel). 2018 Jul 20;9(7):366. doi: 10.3390/genes9070366. Genes (Basel). 2018. PMID: 30037042 Free PMC article.
References
-
- Abbott, A. 2005. Medics braced for fresh superbug. Nature 436758. - PubMed
-
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 2525-29. - PMC - PubMed
-
- Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 325766-5779. - PMC - PubMed
-
- Bratu, S., D. Landman, D. A. Martin, C. Georgescu, and J. Quale. 2008. Correlation of antimicrobial resistance with beta-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob. Agents Chemother. 522999-3005. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

