Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte
- PMID: 18923650
- PMCID: PMC2566589
- DOI: 10.1371/journal.pone.0003408
Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte
Abstract
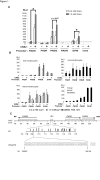
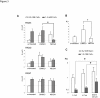
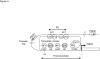
Transcription control at a distance is a critical mechanism, particularly for contiguous genes. The peptidylarginine deiminases (PADs) catalyse the conversion of protein-bound arginine into citrulline (deimination), a critical reaction in the pathophysiology of multiple sclerosis, Alzheimer's disease and rheumatoid arthritis, and in the metabolism of the major epidermal barrier protein filaggrin, a strong predisposing factor for atopic dermatitis. PADs are encoded by 5 clustered PADI genes (1p35-6). Unclear are the mechanisms controlling the expression of the gene PADI3 encoding the PAD3 isoform, a strong candidate for the deimination of filaggrin in the terminally differentiating epidermal keratinocyte. We describe the first PAD Intergenic Enhancer (PIE), an evolutionary conserved non coding segment located 86-kb from the PADI3 promoter. PIE is a strong enhancer of the PADI3 promoter in Ca2+-differentiated epidermal keratinocytes, and requires bound AP-1 factors, namely c-Jun and c-Fos. As compared to proliferative keratinocytes, calcium stimulation specifically associates with increased local DNase I hypersensitivity around PIE, and increased physical proximity of PIE and PADI3 as assessed by Chromosome Conformation Capture. The specific AP-1 inhibitor nordihydroguaiaretic acid suppresses the calcium-induced increase of PADI3 mRNA levels in keratinocytes. Our findings pave the way to the exploration of deimination control during tumorigenesis and wound healing, two conditions for which AP-1 factors are critical, and disclose that long-range transcription control has a role in the regulation of the gene PADI3. Since invalidation of distant regulators causes a variety of human diseases, PIE results to be a plausible candidate in association studies on deimination-related disorders or atopic disease.
Conflict of interest statement
Figures



Similar articles
-
Long-range enhancer differentially regulated by c-Jun and JunD controls peptidylarginine deiminase-3 gene in keratinocytes.J Mol Biol. 2008 Dec 31;384(5):1048-57. doi: 10.1016/j.jmb.2008.10.019. Epub 2008 Oct 15. J Mol Biol. 2008. PMID: 18952102
-
An intronic enhancer driven by NF-κB contributes to transcriptional regulation of peptidylarginine deiminase type I gene in human keratinocytes.J Invest Dermatol. 2010 Nov;130(11):2543-52. doi: 10.1038/jid.2010.179. Epub 2010 Jul 1. J Invest Dermatol. 2010. PMID: 20596086
-
NF-Y and Sp1/Sp3 are involved in the transcriptional regulation of the peptidylarginine deiminase type III gene (PADI3) in human keratinocytes.Biochem J. 2006 Aug 1;397(3):449-59. doi: 10.1042/BJ20051939. Biochem J. 2006. PMID: 16671893 Free PMC article.
-
Transcriptional regulation of peptidylarginine deiminase expression in human keratinocytes.J Dermatol Sci. 2009 Jan;53(1):2-9. doi: 10.1016/j.jdermsci.2008.09.009. Epub 2008 Nov 11. J Dermatol Sci. 2009. PMID: 19004619 Review.
-
Deimination in epidermal barrier and hair formation.Philos Trans R Soc Lond B Biol Sci. 2023 Nov 20;378(1890):20220245. doi: 10.1098/rstb.2022.0245. Epub 2023 Oct 2. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 37778378 Free PMC article. Review.
Cited by
-
High-Resolution Mapping of Multiway Enhancer-Promoter Interactions Regulating Pathogen Detection.Mol Cell. 2020 Oct 15;80(2):359-373.e8. doi: 10.1016/j.molcel.2020.09.005. Epub 2020 Sep 28. Mol Cell. 2020. PMID: 32991830 Free PMC article.
-
Peptidyl Arginine Deiminases in Chronic Diseases: A Focus on Rheumatoid Arthritis and Interstitial Lung Disease.Cells. 2023 Dec 13;12(24):2829. doi: 10.3390/cells12242829. Cells. 2023. PMID: 38132149 Free PMC article. Review.
-
Epstein-Barr virus inactivates the transcriptome and disrupts the chromatin architecture of its host cell in the first phase of lytic reactivation.Nucleic Acids Res. 2021 Apr 6;49(6):3217-3241. doi: 10.1093/nar/gkab099. Nucleic Acids Res. 2021. PMID: 33675667 Free PMC article.
-
Identification of a noncoding RNA‑mediated gene pair‑based regulatory module in Alzheimer's disease.Mol Med Rep. 2018 Aug;18(2):2164-2170. doi: 10.3892/mmr.2018.9190. Epub 2018 Jun 19. Mol Med Rep. 2018. PMID: 29956760 Free PMC article.
-
Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development.Mol Cell. 2017 Sep 21;67(6):1037-1048.e6. doi: 10.1016/j.molcel.2017.08.006. Epub 2017 Sep 7. Mol Cell. 2017. PMID: 28890333 Free PMC article.
References
-
- Chavanas S, Mechin MC, Takahara H, Kawada A, Nachat R, et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene. 2004;330:19–27. - PubMed
-
- Liu GY, Liao YF, Chang WH, Liu CC, Hsieh MC, et al. Overexpression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11:183–196. - PubMed
-
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

