Target identification for CNS diseases by transcriptional profiling
- PMID: 18923405
- PMCID: PMC2675576
- DOI: 10.1038/npp.2008.172
Target identification for CNS diseases by transcriptional profiling
Abstract
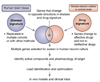
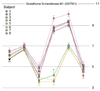
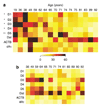
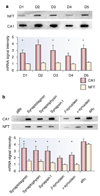
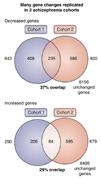
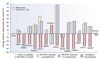
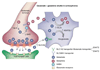
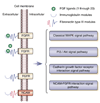
Gene expression changes in neuropsychiatric and neurodegenerative disorders, and gene responses to therapeutic drugs, provide new ways to identify central nervous system (CNS) targets for drug discovery. This review summarizes gene and pathway targets replicated in expression profiling of human postmortem brain, animal models, and cell culture studies. Analysis of isolated human neurons implicates targets for Alzheimer's disease and the cognitive decline associated with normal aging and mild cognitive impairment. In addition to tau, amyloid-beta precursor protein, and amyloid-beta peptides (Abeta), these targets include all three high-affinity neurotrophin receptors and the fibroblast growth factor (FGF) system, synapse markers, glutamate receptors (GluRs) and transporters, and dopamine (DA) receptors, particularly the D2 subtype. Gene-based candidates for Parkinson's disease (PD) include the ubiquitin-proteosome system, scavengers of reactive oxygen species, brain-derived neurotrophic factor (BDNF), its receptor, TrkB, and downstream target early growth response 1, Nurr-1, and signaling through protein kinase C and RAS pathways. Increasing variability and decreases in brain mRNA production from middle age to old age suggest that cognitive impairments during normal aging may be addressed by drugs that restore antioxidant, DNA repair, and synaptic functions including those of DA to levels of younger adults. Studies in schizophrenia identify robust decreases in genes for GABA function, including glutamic acid decarboxylase, HINT1, glutamate transport and GluRs, BDNF and TrkB, numerous 14-3-3 protein family members, and decreases in genes for CNS synaptic and metabolic functions, particularly glycolysis and ATP generation. Many of these metabolic genes are increased by insulin and muscarinic agonism, both of which are therapeutic in psychosis. Differential genomic signals are relatively sparse in bipolar disorder, but include deficiencies in the expression of 14-3-3 protein members, implicating these chaperone proteins and the neurotransmitter pathways they support as possible drug targets. Brains from persons with major depressive disorder reveal decreased expression for genes in glutamate transport and metabolism, neurotrophic signaling (eg, FGF, BDNF and VGF), and MAP kinase pathways. Increases in these pathways in the brains of animals exposed to electroconvulsive shock and antidepressant treatments identify neurotrophic and angiogenic growth factors and second messenger stimulation as therapeutic approaches for the treatment of depression.
Figures



Similar articles
-
Development of allosteric modulators of GPCRs for treatment of CNS disorders.Neurobiol Dis. 2014 Jan;61:55-71. doi: 10.1016/j.nbd.2013.09.013. Epub 2013 Sep 27. Neurobiol Dis. 2014. PMID: 24076101 Free PMC article. Review.
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
-
Neurotrophin Crosstalk in the Etiology and Treatment of Neuropsychiatric and Neurodegenerative Disease.Front Mol Neurosci. 2022 Jul 15;15:932497. doi: 10.3389/fnmol.2022.932497. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35909451 Free PMC article. Review.
-
Dysregulation of TrkB Receptors and BDNF Function by Amyloid-β Peptide is Mediated by Calpain.Cereb Cortex. 2015 Sep;25(9):3107-21. doi: 10.1093/cercor/bhu105. Epub 2014 May 23. Cereb Cortex. 2015. PMID: 24860020
-
Brain-derived neurotrophic factor as a drug target for CNS disorders.Expert Opin Ther Targets. 2004 Oct;8(5):391-9. doi: 10.1517/14728222.8.5.391. Expert Opin Ther Targets. 2004. PMID: 15469390 Review.
Cited by
-
Differential expression of transcriptional regulatory units in the prefrontal cortex of patients with bipolar disorder: potential role of early growth response gene 3.Transl Psychiatry. 2016 May 10;6(5):e805. doi: 10.1038/tp.2016.78. Transl Psychiatry. 2016. PMID: 27163206 Free PMC article.
-
Blood-based biomarkers of Alzheimer's disease: challenging but feasible.Biomark Med. 2010 Feb;4(1):65-79. doi: 10.2217/bmm.09.84. Biomark Med. 2010. PMID: 20387303 Free PMC article. Review.
-
Rapid dopaminergic modulation of the fish hypothalamic transcriptome and proteome.PLoS One. 2010 Aug 20;5(8):e12338. doi: 10.1371/journal.pone.0012338. PLoS One. 2010. PMID: 20808832 Free PMC article.
-
Molecular signatures in post-mortem brain tissue of younger individuals at high risk for Alzheimer's disease as based on APOE genotype.Mol Psychiatry. 2011 Aug;16(8):836-47. doi: 10.1038/mp.2010.57. Epub 2010 May 18. Mol Psychiatry. 2011. PMID: 20479757 Free PMC article.
-
Improving reliability and absolute quantification of human brain microarray data by filtering and scaling probes using RNA-Seq.BMC Genomics. 2014 Feb 24;15(1):154. doi: 10.1186/1471-2164-15-154. BMC Genomics. 2014. PMID: 24564186 Free PMC article.
References
-
- Abe T, Tohgi H, Isobe C, Murata T, Sato C. Remarkable increase in the concentration of 8-hydroxyguanosine in cerebrospinal fluid from patients with Alzheimer’s disease. J Neurosci Res. 2002;70:447–450. - PubMed
-
- ABI. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. Foster City, CA: Applied Biosystems Inc.; 2004. pp. 1–60. Product Guide.
-
- Addington AM, Gornick M, Duckworth J, Sporn A, Gogtay N, Bobb A, et al. GAD1 (2q31.1) which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol Psychiatry. 2005;10:581–588. - PubMed
-
- Agarwal-Mawal A, Qureshi HY, Cafferty PW, Yuan Z, Han V, Lin R, et al. 14-3-3 connects glycogen synthase kinase-3 beta to tau within a brain microtubule-associated tau phosphorylation complex. J Biol Chem. 2003;278:12722–12728. - PubMed
-
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG014449-07/AG/NIA NIH HHS/United States
- P01 AG014449-010003/AG/NIA NIH HHS/United States
- P01 AG017617-090008/AG/NIA NIH HHS/United States
- R21 MH074307-01/MH/NIMH NIH HHS/United States
- P01 AG014449-11A1/AG/NIA NIH HHS/United States
- P01 AG014449-05/AG/NIA NIH HHS/United States
- P01 AG014449-020003/AG/NIA NIH HHS/United States
- P01 NS048447-050001/NS/NINDS NIH HHS/United States
- P01 AG014449-08/AG/NIA NIH HHS/United States
- P01 AG017617-070008/AG/NIA NIH HHS/United States
- AG17617/AG/NIA NIH HHS/United States
- R01 AG043375/AG/NIA NIH HHS/United States
- P01 NS048447-01A20001/NS/NINDS NIH HHS/United States
- AG14449/AG/NIA NIH HHS/United States
- P01 AG017617-080008/AG/NIA NIH HHS/United States
- P01 AG014449-06/AG/NIA NIH HHS/United States
- R21 MH074307-02/MH/NIMH NIH HHS/United States
- P01 NS048447-030001/NS/NINDS NIH HHS/United States
- P01 AG014449-09/AG/NIA NIH HHS/United States
- NS48447/NS/NINDS NIH HHS/United States
- P01 AG014449/AG/NIA NIH HHS/United States
- P01 AG017617-060008/AG/NIA NIH HHS/United States
- R01 MH085801/MH/NIMH NIH HHS/United States
- P01 NS048447-040001/NS/NINDS NIH HHS/United States
- P01 NS048447/NS/NINDS NIH HHS/United States
- P01 AG014449-10/AG/NIA NIH HHS/United States
- P01 AG014449-03/AG/NIA NIH HHS/United States
- R21 MH074307/MH/NIMH NIH HHS/United States
- P01 NS048447-020001/NS/NINDS NIH HHS/United States
- P01 AG017617/AG/NIA NIH HHS/United States
- P01 AG014449-04/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

