Genome-wide association studies: potential next steps on a genetic journey
- PMID: 18852205
- PMCID: PMC2782356
- DOI: 10.1093/hmg/ddn289
Genome-wide association studies: potential next steps on a genetic journey
Abstract
Genome-wide association studies have successfully identified numerous loci at which common variants influence disease risk or quantitative traits. Despite these successes, the variants identified by these studies have generally explained only a small fraction of the heritable component of disease risk, and have not pinpointed with certainty the causal variant(s) at the associated loci. Furthermore, the mechanisms of action by which associated loci influence disease or quantitative phenotypes are often unclear, because we do not know through which gene(s) the associated variants exert their effects or because these gene(s) are of unknown function or have no clear connection to known disease biology. Thus, the initial set of genome-wide association studies serve as a starting point for future genetic and functional studies. We outline possible next steps that may help accelerate progress from genetic studies to the biological knowledge that can guide the development of predictive, preventive, or therapeutic measures.
Figures
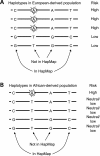

Similar articles
-
Rare-variant genome-wide association studies: a new frontier in genetic analysis of complex traits.Pharmacogenomics. 2013 Mar;14(4):413-24. doi: 10.2217/pgs.13.36. Pharmacogenomics. 2013. PMID: 23438888 Review.
-
Genome-wide association studies: results from the first few years and potential implications for clinical medicine.Annu Rev Med. 2011;62:11-24. doi: 10.1146/annurev.med.091708.162036. Annu Rev Med. 2011. PMID: 21226609 Review.
-
Genome-wide association studies of human growth traits.Nestle Nutr Inst Workshop Ser. 2013;71:29-38. doi: 10.1159/000342541. Epub 2013 Jan 22. Nestle Nutr Inst Workshop Ser. 2013. PMID: 23502136 Review.
-
Autism genetics: emerging data from genome-wide copy-number and single nucleotide polymorphism scans.Expert Rev Mol Diagn. 2009 Nov;9(8):795-803. doi: 10.1586/erm.09.59. Expert Rev Mol Diagn. 2009. PMID: 19895225 Review.
-
Rare variants and cardiovascular disease.Brief Funct Genomics. 2014 Sep;13(5):384-91. doi: 10.1093/bfgp/elu010. Epub 2014 Apr 26. Brief Funct Genomics. 2014. PMID: 24771349 Review.
Cited by
-
Beyond the fourth wave of genome-wide obesity association studies.Nutr Diabetes. 2012 Jul 30;2(7):e37. doi: 10.1038/nutd.2012.9. Nutr Diabetes. 2012. PMID: 23168490 Free PMC article.
-
Individualized risk for statin-induced myopathy: current knowledge, emerging challenges and potential solutions.Pharmacogenomics. 2012 Apr;13(5):579-94. doi: 10.2217/pgs.12.11. Pharmacogenomics. 2012. PMID: 22462750 Free PMC article. Review.
-
Design of association studies with pooled or un-pooled next-generation sequencing data.Genet Epidemiol. 2010 Jul;34(5):479-91. doi: 10.1002/gepi.20501. Genet Epidemiol. 2010. PMID: 20552648 Free PMC article.
-
The psychiatric GWAS consortium: big science comes to psychiatry.Neuron. 2010 Oct 21;68(2):182-6. doi: 10.1016/j.neuron.2010.10.003. Neuron. 2010. PMID: 20955924 Free PMC article.
-
Zebrafish genetic models for arrhythmia.Prog Biophys Mol Biol. 2008 Oct-Nov;98(2-3):301-8. doi: 10.1016/j.pbiomolbio.2009.01.011. Epub 2009 Jan 31. Prog Biophys Mol Biol. 2008. PMID: 19351520 Free PMC article. Review.
References
-
- Easton D. Genome-wide association studies in cancer. Hum. Mol. Genet. 2008 (this issue) - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

