Analysis of gene regulatory networks in the mammalian circadian rhythm
- PMID: 18846204
- PMCID: PMC2543109
- DOI: 10.1371/journal.pcbi.1000193
Analysis of gene regulatory networks in the mammalian circadian rhythm
Abstract
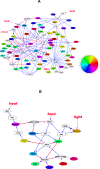
Circadian rhythm is fundamental in regulating a wide range of cellular, metabolic, physiological, and behavioral activities in mammals. Although a small number of key circadian genes have been identified through extensive molecular and genetic studies in the past, the existence of other key circadian genes and how they drive the genomewide circadian oscillation of gene expression in different tissues still remains unknown. Here we try to address these questions by integrating all available circadian microarray data in mammals. We identified 41 common circadian genes that showed circadian oscillation in a wide range of mouse tissues with a remarkable consistency of circadian phases across tissues. Comparisons across mouse, rat, rhesus macaque, and human showed that the circadian phases of known key circadian genes were delayed for 4-5 hours in rat compared to mouse and 8-12 hours in macaque and human compared to mouse. A systematic gene regulatory network for the mouse circadian rhythm was constructed after incorporating promoter analysis and transcription factor knockout or mutant microarray data. We observed the significant association of cis-regulatory elements: EBOX, DBOX, RRE, and HSE with the different phases of circadian oscillating genes. The analysis of the network structure revealed the paths through which light, food, and heat can entrain the circadian clock and identified that NR3C1 and FKBP/HSP90 complexes are central to the control of circadian genes through diverse environmental signals. Our study improves our understanding of the structure, design principle, and evolution of gene regulatory networks involved in the mammalian circadian rhythm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A network biology study on circadian rhythm by integrating various omics data.OMICS. 2009 Aug;13(4):313-24. doi: 10.1089/omi.2009.0040. OMICS. 2009. PMID: 19645592
-
Transcriptional oscillation of canonical clock genes in mouse peripheral tissues.BMC Mol Biol. 2004 Oct 9;5:18. doi: 10.1186/1471-2199-5-18. BMC Mol Biol. 2004. PMID: 15473909 Free PMC article.
-
Promoter analysis of Mammalian clock controlled genes.Genome Inform. 2007;18:65-74. Genome Inform. 2007. PMID: 18546475
-
Chronobiology of micturition: putative role of the circadian clock.J Urol. 2013 Sep;190(3):843-9. doi: 10.1016/j.juro.2013.02.024. Epub 2013 Feb 18. J Urol. 2013. PMID: 23429068 Review.
-
Molecular genetics of circadian rhythms in mammals.Annu Rev Neurosci. 2000;23:713-42. doi: 10.1146/annurev.neuro.23.1.713. Annu Rev Neurosci. 2000. PMID: 10845079 Review.
Cited by
-
Comparative transcriptomic rhythms in the mouse and human prefrontal cortex.bioRxiv [Preprint]. 2024 Sep 26:2024.09.26.615154. doi: 10.1101/2024.09.26.615154. bioRxiv. 2024. PMID: 39386590 Free PMC article. Preprint.
-
Modulation of Neuronal Excitability and Plasticity by BHLHE41 Conveys Lithium Non-Responsiveness.bioRxiv [Preprint]. 2024 Jul 25:2024.07.25.605130. doi: 10.1101/2024.07.25.605130. bioRxiv. 2024. PMID: 39372797 Free PMC article. Preprint.
-
Comparative rhythmic transcriptome profiling of human and mouse striatal subregions.Neuropsychopharmacology. 2024 Apr;49(5):796-805. doi: 10.1038/s41386-023-01788-w. Epub 2024 Jan 5. Neuropsychopharmacology. 2024. PMID: 38182777
-
Astrocytic insulin receptor controls circadian behavior via dopamine signaling in a sexually dimorphic manner.Nat Commun. 2023 Dec 9;14(1):8175. doi: 10.1038/s41467-023-44039-8. Nat Commun. 2023. PMID: 38071352 Free PMC article.
-
Circadian clock regulator Bmal1 gates axon regeneration via Tet3 epigenetics in mouse sensory neurons.Nat Commun. 2023 Aug 24;14(1):5165. doi: 10.1038/s41467-023-40816-7. Nat Commun. 2023. PMID: 37620297 Free PMC article.
References
-
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. - PubMed
-
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. - PubMed
-
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. - PubMed
-
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

