Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1
- PMID: 18840615
- PMCID: PMC2662239
- DOI: 10.1074/jbc.M803925200
Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1
Abstract
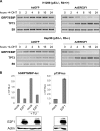
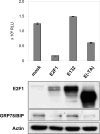
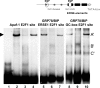
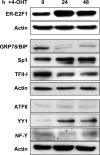
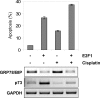
The endoplasmic reticulum chaperone GRP78/BIP plays a central role in the prosurvival machinery, and its enhanced expression has been implicated in drug resistance, carcinogenesis, and metastasis. E2F1, as part of an antitumor safeguard mechanism, promotes apoptosis regardless of functional p53. Using cells that are defective in p53, we show that E2F1 represses GRP78/BIP at the transcriptional level, and this requires its DNA binding domain. Analysis of human GRP78/BIP promoter reporter constructs revealed that the region between -371 and -109 of the proximal promoter contains major E2F1-responsive elements. Toward understanding the underlying mechanism of this regulation, we performed chromatin immunoprecipitation and gel shift assays, demonstrating that E2F1 directly binds to GC-rich regions in the distal GC-box and endoplasmic reticulum stress response element -126 by interfering with the binding of positive regulatory proteins Sp1 and TFII-I of the ER stress response element-binding factor complex. We further show that TFII-I, which is required for optimal stress induction of GRP78/BIP, is suppressed by E2F1 on the protein level. Finally, our studies suggest a molecular link between the inhibition of GRP78/BIP and E2F1-mediated chemosensitization of tumor cells, underscoring its relevance for cancer treatment. Together, the data provide a new mechanism for the incompletely understood tumor suppressor function of E2F1.
Figures



Similar articles
-
Transcriptional regulation of the Grp78 promoter by endoplasmic reticulum stress: role of TFII-I and its tyrosine phosphorylation.J Biol Chem. 2005 Apr 29;280(17):16821-8. doi: 10.1074/jbc.M413753200. Epub 2005 Jan 21. J Biol Chem. 2005. PMID: 15664986
-
Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers.Mol Cell Biol. 2005 Jun;25(11):4529-40. doi: 10.1128/MCB.25.11.4529-4540.2005. Mol Cell Biol. 2005. PMID: 15899857 Free PMC article.
-
In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4.J Biol Chem. 2006 Mar 31;281(13):8877-87. doi: 10.1074/jbc.M505784200. Epub 2006 Feb 1. J Biol Chem. 2006. PMID: 16452489
-
The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends.J Biol Chem. 2019 Feb 8;294(6):2098-2108. doi: 10.1074/jbc.REV118.002804. Epub 2018 Dec 18. J Biol Chem. 2019. PMID: 30563838 Free PMC article. Review.
-
GRP78 in lung cancer.J Transl Med. 2021 Mar 21;19(1):118. doi: 10.1186/s12967-021-02786-6. J Transl Med. 2021. PMID: 33743739 Free PMC article. Review.
Cited by
-
Ribosomal protein L5 (RPL5)/ E2F transcription factor 1 (E2F1) signaling suppresses breast cancer progression via regulating endoplasmic reticulum stress and autophagy.Bioengineered. 2022 Apr;13(4):8076-8086. doi: 10.1080/21655979.2022.2052672. Bioengineered. 2022. PMID: 35293275 Free PMC article.
-
Targeting Rb inactivation in cancers by synthetic lethality.Am J Cancer Res. 2011 Jun 30;1(6):773-86. Am J Cancer Res. 2011. PMID: 21814623 Free PMC article.
-
E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation.EMBO Rep. 2014 Dec;15(12):1315-29. doi: 10.15252/embr.201439392. Epub 2014 Oct 23. EMBO Rep. 2014. PMID: 25341426 Free PMC article.
-
LINC00629 protects osteosarcoma cell from ER stress-induced apoptosis and facilitates tumour progression by elevating KLF4 stability.J Exp Clin Cancer Res. 2022 Dec 20;41(1):354. doi: 10.1186/s13046-022-02569-x. J Exp Clin Cancer Res. 2022. PMID: 36539799 Free PMC article.
-
GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria.Cell Death Differ. 2011 May;18(5):874-86. doi: 10.1038/cdd.2010.153. Epub 2010 Dec 3. Cell Death Differ. 2011. PMID: 21127500 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

