Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis
- PMID: 18835819
- PMCID: PMC2586264
- DOI: 10.1074/jbc.M803312200
Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis
Abstract
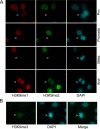
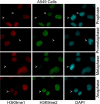
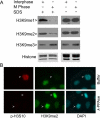
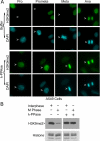
Post-translational modifications of histones play a critical role in regulating genome structures and integrity. We have focused on the regulatory relationship between covalent modifications of histone H3 lysine 9 (H3K9) and H3S10 during the cell cycle. Immunofluorescence microscopy revealed that H3S10 phosphorylation in HeLa, A549, and HCT116 cells was high during prophase, prometaphase, and metaphase, whereas H3K9 monomethylation (H3K9me1) and dimethylation (H3K9me2), but not H3K9 trimethylation (H3K9me3), were significantly suppressed. When H3S10 phosphorylation started to diminish during anaphase, H3K9me1 and H3K9me2 signals reemerged. Western blot analyses confirmed that mitotic histones, extracted in an SDS-containing buffer, had little H3K9me1 and H3K9me2 signals but abundant H3K9me3 signals. However, when mitotic histones were extracted in the same buffer without SDS, the difference in H3K9me1 and H3K9me2 signals between interphase and mitotic cells disappeared. Removal of H3S10 phosphorylation by pretreatment with lambda-phosphatase unmasked mitotic H3K9me1 and H3K9me2 signals detected by both fluorescence microscopy and Western blotting. Further, H3S10 phosphorylation completely blocked methylation of H3K9 but not demethylation of the same residue in vitro. Given that several conserved motifs consisting of a Lys residue immediately followed by a Ser residue are present in histone tails, our studies reveal a potential new mechanism by which phosphorylation not only regulates selective access of methylated lysines by cellular factors but also serves to preserve methylation patterns and epigenetic programs during cell division.
Figures


Similar articles
-
Regulation of chromatin structure by histone H3S10 phosphorylation.Chromosome Res. 2006;14(4):393-404. doi: 10.1007/s10577-006-1063-4. Chromosome Res. 2006. PMID: 16821135 Review.
-
Phosphorylation of serine-10 of histone H3 shields modified lysine-9 selectively during mitosis.Genes Cells. 2010 Mar;15(3):181-92. doi: 10.1111/j.1365-2443.2009.01375.x. Epub 2010 Jan 13. Genes Cells. 2010. PMID: 20070858
-
Dynamic regulation of histone modifications in Xenopus oocytes through histone exchange.Mol Cell Biol. 2006 Sep;26(18):6890-901. doi: 10.1128/MCB.00948-06. Mol Cell Biol. 2006. PMID: 16943430 Free PMC article.
-
Domain requirements of the JIL-1 tandem kinase for histone H3 serine 10 phosphorylation and chromatin remodeling in vivo.J Biol Chem. 2013 Jul 5;288(27):19441-9. doi: 10.1074/jbc.M113.464271. Epub 2013 May 30. J Biol Chem. 2013. PMID: 23723094 Free PMC article.
-
Regulation and function of H3K9 methylation.Subcell Biochem. 2007;41:337-50. Subcell Biochem. 2007. PMID: 17484135 Review.
Cited by
-
Epigenetic histone modifications in a clinically relevant rat model of chronic ethanol-binge-mediated liver injury.Hepatol Int. 2014 Sep;8 Suppl 2:421-30. doi: 10.1007/s12072-014-9546-4. Epub 2014 Jun 18. Hepatol Int. 2014. PMID: 26201320
-
Examining histone posttranslational modification patterns by high-resolution mass spectrometry.Methods Enzymol. 2012;512:3-28. doi: 10.1016/B978-0-12-391940-3.00001-9. Methods Enzymol. 2012. PMID: 22910200 Free PMC article.
-
Histone regulation in the CNS: basic principles of epigenetic plasticity.Neuropsychopharmacology. 2013 Jan;38(1):3-22. doi: 10.1038/npp.2012.124. Epub 2012 Jul 25. Neuropsychopharmacology. 2013. PMID: 22828751 Free PMC article. Review.
-
Quantifying epigenetic modulation of nucleosome breathing by high-throughput AFM imaging.Biophys J. 2022 Mar 1;121(5):841-851. doi: 10.1016/j.bpj.2022.01.014. Epub 2022 Jan 20. Biophys J. 2022. PMID: 35065917 Free PMC article.
-
A computational model for histone mark propagation reproduces the distribution of heterochromatin in different human cell types.PLoS One. 2013 Sep 19;8(9):e73818. doi: 10.1371/journal.pone.0073818. eCollection 2013. PLoS One. 2013. PMID: 24069233 Free PMC article.
References
-
- Rice, K. L., Hormaeche, I., and Licht, J. D. (2007) Oncogene 26 6697-6714 - PubMed
-
- Shilatifard, A. (2006) Annu. Rev. Biochem. 75 243-269 - PubMed
-
- Bode, A. M., and Dong, Z. (2005) Science's STKE 2005 re4 - PubMed
-
- Nowak, S. J., and Corces, V. G. (2004) Trends Genet. 20 214-220 - PubMed
-
- Prigent, C., and Dimitrov, S. (2003) J. Cell Sci. 116 3677-3685 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

