Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator
- PMID: 18828672
- PMCID: PMC2553839
- DOI: 10.1371/journal.pbio.0060233
Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator
Abstract
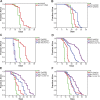
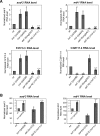
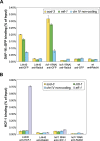
The transcription factor DAF-16/forkhead box O (FOXO) is a critical longevity determinant in diverse organisms, however the molecular basis of how its transcriptional activity is regulated remains largely unknown. We report that the Caenorhabditis elegans homolog of host cell factor 1 (HCF-1) represents a new longevity modulator and functions as a negative regulator of DAF-16. In C. elegans, hcf-1 inactivation caused a daf-16-dependent lifespan extension of up to 40% and heightened resistance to specific stress stimuli. HCF-1 showed ubiquitous nuclear localization and physically associated with DAF-16. Furthermore, loss of hcf-1 resulted in elevated DAF-16 recruitment to the promoters of its target genes and altered expression of a subset of DAF-16-regulated genes. We propose that HCF-1 modulates C. elegans longevity and stress response by forming a complex with DAF-16 and limiting a fraction of DAF-16 from accessing its target gene promoters, and thereby regulates DAF-16-mediated transcription of selective target genes. As HCF-1 is highly conserved, our findings have important implications for aging and FOXO regulation in mammals.
Conflict of interest statement
Figures



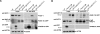

Similar articles
-
The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO.PLoS Genet. 2011 Sep;7(9):e1002235. doi: 10.1371/journal.pgen.1002235. Epub 2011 Sep 1. PLoS Genet. 2011. PMID: 21909281 Free PMC article.
-
SMK-1, an essential regulator of DAF-16-mediated longevity.Cell. 2006 Mar 10;124(5):1039-53. doi: 10.1016/j.cell.2005.12.042. Cell. 2006. PMID: 16530049
-
Host cell factor 1 inhibits SKN-1 to modulate oxidative stress responses in Caenorhabditis elegans.Aging Cell. 2012 Aug;11(4):717-21. doi: 10.1111/j.1474-9726.2012.00831.x. Epub 2012 Jun 4. Aging Cell. 2012. PMID: 22568582 Free PMC article.
-
The search for DAF-16/FOXO transcriptional targets: approaches and discoveries.Exp Gerontol. 2006 Oct;41(10):910-21. doi: 10.1016/j.exger.2006.06.040. Epub 2006 Aug 24. Exp Gerontol. 2006. PMID: 16934425 Review.
-
DAF-16: FOXO in the Context of C. elegans.Curr Top Dev Biol. 2018;127:1-21. doi: 10.1016/bs.ctdb.2017.11.007. Epub 2018 Feb 2. Curr Top Dev Biol. 2018. PMID: 29433733 Review.
Cited by
-
Pathogenesis of the Candida parapsilosis Complex in the Model Host Caenorhabditis elegans.Genes (Basel). 2018 Aug 8;9(8):401. doi: 10.3390/genes9080401. Genes (Basel). 2018. PMID: 30096852 Free PMC article.
-
An Intestinal Symbiotic Bacterial Strain of Oscheius chongmingensis Modulates Host Viability at Both Global and Post-Transcriptional Levels.Int J Mol Sci. 2022 Nov 24;23(23):14692. doi: 10.3390/ijms232314692. Int J Mol Sci. 2022. PMID: 36499019 Free PMC article.
-
MSP hormonal control of the oocyte MAP kinase cascade and reactive oxygen species signaling.Dev Biol. 2010 Jun 1;342(1):96-107. doi: 10.1016/j.ydbio.2010.03.026. Epub 2010 Apr 7. Dev Biol. 2010. PMID: 20380830 Free PMC article.
-
daf-16/FOXO blocks adult cell fate in Caenorhabditis elegans dauer larvae via lin-41/TRIM71.PLoS Genet. 2021 Nov 15;17(11):e1009881. doi: 10.1371/journal.pgen.1009881. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34780472 Free PMC article.
-
TATN-1 mutations reveal a novel role for tyrosine as a metabolic signal that influences developmental decisions and longevity in Caenorhabditis elegans.PLoS Genet. 2013;9(12):e1004020. doi: 10.1371/journal.pgen.1004020. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24385923 Free PMC article.
References
-
- Antebi A. Genetics of aging in Caenorhabditis elegans . PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. - DOI - PMC - PubMed
-
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. - PubMed
-
- Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32:180–188. - PubMed
-
- Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. - DOI - PMC - PubMed
-
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

