Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses
- PMID: 18818732
- PMCID: PMC2533123
- DOI: 10.1371/journal.ppat.1000161
Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses
Abstract
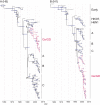
The highly pathogenic avian influenza (HPAI) H5N1 virus lineage has undergone extensive genetic reassortment with viruses from different sources to produce numerous H5N1 genotypes, and also developed into multiple genetically distinct sublineages in China. From there, the virus has spread to over 60 countries. The ecological success of this virus in diverse species of both poultry and wild birds with frequent introduction to humans suggests that it is a likely source of the next human pandemic. Therefore, the evolutionary and ecological characteristics of its emergence from wild birds into poultry are of considerable interest. Here, we apply the latest analytical techniques to infer the early evolutionary dynamics of H5N1 virus in the population from which it emerged (wild birds and domestic poultry). By estimating the time of most recent common ancestors of each gene segment, we show that the H5N1 prototype virus was likely introduced from wild birds into poultry as a non-reassortant low pathogenic avian influenza H5N1 virus and was not generated by reassortment in poultry. In contrast, more recent H5N1 genotypes were generated locally in aquatic poultry after the prototype virus (A/goose/Guangdong/1/96) introduction occurred, i.e., they were not a result of additional emergence from wild birds. We show that the H5N1 virus was introduced into Indonesia and Vietnam 3-6 months prior to detection of the first outbreaks in those countries. Population dynamics analyses revealed a rapid increase in the genetic diversity of A/goose/Guangdong/1/96 lineage viruses from mid-1999 to early 2000. Our results suggest that the transmission of reassortant viruses through the mixed poultry population in farms and markets in China has selected HPAI H5N1 viruses that are well adapted to multiple hosts and reduced the interspecies transmission barrier of those viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses.J Virol. 2007 Jul;81(14):7529-39. doi: 10.1128/JVI.00327-07. Epub 2007 May 16. J Virol. 2007. PMID: 17507485 Free PMC article.
-
Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam.Virology. 2006 Jul 5;350(2):258-68. doi: 10.1016/j.virol.2006.03.048. Epub 2006 May 19. Virology. 2006. PMID: 16713612
-
Genetic analysis of avian influenza A viruses isolated from domestic waterfowl in live-bird markets of Hanoi, Vietnam, preceding fatal H5N1 human infections in 2004.Arch Virol. 2009;154(8):1249-61. doi: 10.1007/s00705-009-0429-2. Epub 2009 Jul 4. Arch Virol. 2009. PMID: 19578928
-
Origin and evolution of highly pathogenic H5N1 avian influenza in Asia.Vet Rec. 2005 Aug 6;157(6):159-64. doi: 10.1136/vr.157.6.159. Vet Rec. 2005. PMID: 16085721 Review.
-
H5N1 avian influenza in China.Sci China C Life Sci. 2009 May;52(5):419-27. doi: 10.1007/s11427-009-0068-6. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471864 Review.
Cited by
-
Ecosystem Interactions Underlie the Spread of Avian Influenza A Viruses with Pandemic Potential.PLoS Pathog. 2016 May 11;12(5):e1005620. doi: 10.1371/journal.ppat.1005620. eCollection 2016 May. PLoS Pathog. 2016. PMID: 27166585 Free PMC article.
-
Modelling the progression of pandemic influenza A (H1N1) in Vietnam and the opportunities for reassortment with other influenza viruses.BMC Med. 2009 Sep 3;7:43. doi: 10.1186/1741-7015-7-43. BMC Med. 2009. PMID: 19728864 Free PMC article.
-
Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview.Viruses. 2018 Sep 13;10(9):497. doi: 10.3390/v10090497. Viruses. 2018. PMID: 30217093 Free PMC article. Review.
-
Immunogenicity and Cross-Protective Efficacy Induced by an Inactivated Recombinant Avian Influenza A/H5N1 (Clade 2.3.4.4b) Vaccine against Co-Circulating Influenza A/H5Nx Viruses.Vaccines (Basel). 2023 Aug 22;11(9):1397. doi: 10.3390/vaccines11091397. Vaccines (Basel). 2023. PMID: 37766075 Free PMC article.
-
A(H5N1) Virus Evolution in South East Asia.Viruses. 2009 Dec;1(3):335-61. doi: 10.3390/v1030335. Epub 2009 Oct 6. Viruses. 2009. PMID: 21994553 Free PMC article.
References
-
- Xu X, Subbarao K, Cox NJ, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. - PubMed
-
- Thiry E, Zicola D, Addie H, Egberink K, Hartmann H, et al. Highly pathogenic avian influenza in cats and other carnivores. Vet Microbiol. 2007;122:25–31. - PubMed
-
- World Health Organization. Cumulative number of confirmed human cases of Avian Influenza A (H5N1) reported to WHO (WHO, Geneva). 2008. Available: http://www.who.int/csr/disease/avian_influenza/country/en/. Accessed 11 August 2008.
-
- Li KS, Guan Y, Wang J, Smith GJD, Xu KM, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

