An ontology for Xenopus anatomy and development
- PMID: 18817563
- PMCID: PMC2561031
- DOI: 10.1186/1471-213X-8-92
An ontology for Xenopus anatomy and development
Abstract
Background: The frogs Xenopus laevis and Xenopus (Silurana) tropicalis are model systems that have produced a wealth of genetic, genomic, and developmental information. Xenbase is a model organism database that provides centralized access to this information, including gene function data from high-throughput screens and the scientific literature. A controlled, structured vocabulary for Xenopus anatomy and development is essential for organizing these data.
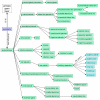
Results: We have constructed a Xenopus anatomical ontology that represents the lineage of tissues and the timing of their development. We have classified many anatomical features in a common framework that has been adopted by several model organism database communities. The ontology is available for download at the Open Biomedical Ontologies Foundry http://obofoundry.org.
Conclusion: The Xenopus Anatomical Ontology will be used to annotate Xenopus gene expression patterns and mutant and morphant phenotypes. Its robust developmental map will enable powerful database searches and data analyses. We encourage community recommendations for updates and improvements to the ontology.
Figures
Similar articles
-
Xenbase: a Xenopus biology and genomics resource.Nucleic Acids Res. 2008 Jan;36(Database issue):D761-7. doi: 10.1093/nar/gkm826. Epub 2007 Nov 4. Nucleic Acids Res. 2008. PMID: 17984085 Free PMC article.
-
Enhanced XAO: the ontology of Xenopus anatomy and development underpins more accurate annotation of gene expression and queries on Xenbase.J Biomed Semantics. 2013 Oct 18;4(1):31. doi: 10.1186/2041-1480-4-31. J Biomed Semantics. 2013. PMID: 24139024 Free PMC article.
-
Xenbase: key features and resources of the Xenopus model organism knowledgebase.Genetics. 2023 May 4;224(1):iyad018. doi: 10.1093/genetics/iyad018. Genetics. 2023. PMID: 36755307 Free PMC article.
-
Xenopus genomic data and browser resources.Dev Biol. 2017 Jun 15;426(2):194-199. doi: 10.1016/j.ydbio.2016.03.030. Epub 2016 Mar 31. Dev Biol. 2017. PMID: 27039265 Free PMC article. Review.
-
Heading in a new direction: implications of the revised fate map for understanding Xenopus laevis development.Dev Biol. 2006 Aug 1;296(1):12-28. doi: 10.1016/j.ydbio.2006.04.447. Epub 2006 Apr 21. Dev Biol. 2006. PMID: 16750823 Review.
Cited by
-
Evaluation of research in biomedical ontologies.Brief Bioinform. 2013 Nov;14(6):696-712. doi: 10.1093/bib/bbs053. Epub 2012 Sep 8. Brief Bioinform. 2013. PMID: 22962340 Free PMC article.
-
Bio-SimVerb and Bio-SimLex: wide-coverage evaluation sets of word similarity in biomedicine.BMC Bioinformatics. 2018 Feb 5;19(1):33. doi: 10.1186/s12859-018-2039-z. BMC Bioinformatics. 2018. PMID: 29402212 Free PMC article.
-
The skin microbiome of Xenopus laevis and the effects of husbandry conditions.Anim Microbiome. 2021 Feb 5;3(1):17. doi: 10.1186/s42523-021-00080-w. Anim Microbiome. 2021. PMID: 33546771 Free PMC article.
-
Probing the Xenopus laevis inner ear transcriptome for biological function.BMC Genomics. 2012 Jun 8;13:225. doi: 10.1186/1471-2164-13-225. BMC Genomics. 2012. PMID: 22676585 Free PMC article.
-
Navigating Xenbase: An Integrated Xenopus Genomics and Gene Expression Database.Methods Mol Biol. 2018;1757:251-305. doi: 10.1007/978-1-4939-7737-6_10. Methods Mol Biol. 2018. PMID: 29761462 Free PMC article.
References
-
- Nieuwkoop PD, Faber J, (Eds) Normal Table of Xenopus laevis (Daudin) New York: Garland Publishing; 1994.
-
- NIBB XDB3 http://xenopus.nibb.ac.jp
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

