Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging
- PMID: 18816868
- PMCID: PMC2579754
- DOI: 10.1002/mrm.21712
Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging
Abstract
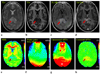
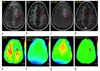
Amide proton transfer (APT) imaging is a type of chemical exchange-dependent saturation transfer (CEST) magnetic resonance imaging (MRI) in which amide protons of endogenous mobile proteins and peptides in tissue are detected. Initial studies have shown promising results for distinguishing tumor from surrounding brain in patients, but these data were hampered by magnetic field inhomogeneity and a low signal-to-noise ratio (SNR). Here a practical six-offset APT data acquisition scheme is presented that, together with a separately acquired CEST spectrum, can provide B(0)-inhomogeneity corrected human brain APT images of sufficient SNR within a clinically relevant time frame. Data from nine brain tumor patients at 3T shows that APT intensities were significantly higher in the tumor core, as assigned by gadolinium-enhancement, than in contralateral normal-appearing white matter (CNAWM) in patients with high-grade tumors. Conversely, APT intensities in tumor were indistinguishable from CNAWM in patients with low-grade tumors. In high-grade tumors, regions of increased APT extended outside of the core into peripheral zones, indicating the potential of this technique for more accurate delineation of the heterogeneous areas of brain cancers.
(c) 2008 Wiley-Liss, Inc.
Figures


Similar articles
-
Whole-brain amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging in glioma patients using low-power steady-state pulsed chemical exchange saturation transfer (CEST) imaging at 7T.J Magn Reson Imaging. 2016 Jul;44(1):41-50. doi: 10.1002/jmri.25108. Epub 2015 Dec 10. J Magn Reson Imaging. 2016. PMID: 26663561 Free PMC article.
-
Amide proton transfer imaging of human brain tumors at 3T.Magn Reson Med. 2006 Sep;56(3):585-92. doi: 10.1002/mrm.20989. Magn Reson Med. 2006. PMID: 16892186 Clinical Trial.
-
Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 Tesla.Magn Reson Med. 2016 Jan;75(1):137-49. doi: 10.1002/mrm.25581. Epub 2015 Mar 5. Magn Reson Med. 2016. PMID: 25753614 Free PMC article.
-
Amide proton transfer imaging of tumors: theory, clinical applications, pitfalls, and future directions.Jpn J Radiol. 2019 Feb;37(2):109-116. doi: 10.1007/s11604-018-0787-3. Epub 2018 Oct 19. Jpn J Radiol. 2019. PMID: 30341472 Review.
-
APT-weighted MRI: Techniques, current neuro applications, and challenging issues.J Magn Reson Imaging. 2019 Aug;50(2):347-364. doi: 10.1002/jmri.26645. Epub 2019 Jan 20. J Magn Reson Imaging. 2019. PMID: 30663162 Free PMC article. Review.
Cited by
-
Adjustment of rotation and saturation effects (AROSE) for CEST imaging.Magn Reson Med. 2024 Mar;91(3):1016-1029. doi: 10.1002/mrm.29938. Epub 2023 Nov 27. Magn Reson Med. 2024. PMID: 38009992
-
Characterizing amide proton transfer imaging in haemorrhage brain lesions using 3T MRI.Eur Radiol. 2017 Apr;27(4):1577-1584. doi: 10.1007/s00330-016-4477-1. Epub 2016 Jul 6. Eur Radiol. 2017. PMID: 27380905 Free PMC article.
-
Comparison of Diffusion Kurtosis Imaging and Amide Proton Transfer Imaging in the Diagnosis and Risk Assessment of Prostate Cancer.Front Oncol. 2021 Apr 15;11:640906. doi: 10.3389/fonc.2021.640906. eCollection 2021. Front Oncol. 2021. PMID: 33937041 Free PMC article.
-
A Brief History and Future Prospects of CEST MRI in Clinical Non-Brain Tumor Imaging.Int J Mol Sci. 2021 Oct 26;22(21):11559. doi: 10.3390/ijms222111559. Int J Mol Sci. 2021. PMID: 34768990 Free PMC article. Review.
-
Relaxation-compensated amide proton transfer (APT) MRI signal intensity is associated with survival and progression in high-grade glioma patients.Eur Radiol. 2019 Sep;29(9):4957-4967. doi: 10.1007/s00330-019-06066-2. Epub 2019 Feb 26. Eur Radiol. 2019. PMID: 30809720
References
-
- Hobbs SK, Shi G, Homer R, Harsh G, Altlas SW, Bednarski MD. Magnetic resonance imaging-guided proteomics of human glioblastoma multiforme. J Magn Reson Imag. 2003;18:530–536. - PubMed
-
- Li J, Zhuang Z, Okamoto H, Vortmeyer AO, Park DM, Furata M, Lee Y-S, Oldfield EH, Zeng W, Weil RJ. Proteomic profiling distinguishes astrocytomas and identifies differential tumor markers. Neurology. 2006;66:733–736. - PubMed
-
- Shen J, Behrens B, Wistuba II, Feng L, Lee JJ, Hong WK, Lotan R. Identification and validation of differences in protein levels in normal, premalignant, and malignant lung cells and tissues using high-throughput Western array and immunohischemistry. Cancer Res. 2006;66:11194–11206. - PubMed
-
- Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, Bell BA, Griffiths JR. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–232. - PubMed
-
- Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

